Abstract
The ability to edit DNA at the nucleotide level using clustered regularly interspaced short palindromic repeats (CRISPR) systems is a relatively new investigative tool that is revolutionizing the analysis of many aspects of human health and disease, including orthopaedic disease. CRISPR, adapted for mammalian cell genome editing from a bacterial defence system, has been shown to be a flexible, programmable, scalable, and easy-to-use gene editing tool. Recent improvements increase the functionality of CRISPR through the engineering of specific elements of CRISPR systems, the discovery of new, naturally occurring CRISPR molecules, and modifications that take CRISPR beyond gene editing to the regulation of gene transcription and the manipulation of RNA. Here, the basics of CRISPR genome editing will be reviewed, including a description of how it has transformed some aspects of molecular musculoskeletal research, and will conclude by speculating what the future holds for the use of CRISPR-related treatments and therapies in clinical orthopaedic practice.
Cite this article: Bone Joint Res 2020;9(7):351–359.
Article focus
-
Applications of clustered regularly interspaced short palindromic repeats (CRISPR) technologies to musculoskeletal research.
Key messages
-
CRISPR systems are in widespread use in musculoskeletal research.
-
Applications include gene ‘knock-in’ and ‘knock-out’ cell models and in small and large vertebrate animal models.
-
Novel applications and adaptations extend the functionality of CRISPR.
Strengths and limitations
-
This study focusses on the growing use of CRISPR technologies in musculoskeletal and orthopaedic applications.
-
This is a rapidly advancing field and information in this review article may become surpassed by new advances.
Introduction
At its simplest, genome editing refers to the ability to permanently alter DNA at the resolution of the nucleotide. Since its discovery and practical application first with transcription activator-like effector nuclease (TALEN) systems1,2 and then with clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9,3,4 DNA editing has become one of the most powerful experimental tools available to biologists. The intellectual origins for genome editing can be traced back to the foundational years of molecular biology where the central dogma of molecular biology, first articulated by Crick in the early 1960s, states that sequential information passes from nucleic acid to protein in one direction. This means that permanent edits made in DNA have the potential to alter protein sequences and, therefore, protein function in predictable ways. While this makes CRISPR a powerful experimental and potentially therapeutic tool, the reality is more interesting. In addition to the simplest cases where genes (and therefore proteins) are altered at the nucleotide levels or ‘knocked-out’ (KO), there exists an almost unlimited range of ‘knock-in’ (KI) DNA alterations that can be made. These include the introduction of fluorescent markers to trace gene activation, the inclusion of specific mutations to model disease, and the reversible regulation of gene transcription for probing developmental processes, to name a few. Here, we will cover the basics of CRISPR-Cas9 editing and describe the latest developments in editing as they relate to orthopaedic biology, including exciting developments that expand beyond mouse models to larger, more translationally relevant, animal model systems. Papers were identified for this review by searching PubMed using terms that combined “CRISPR”, “CRISPR-Cas9”, “genome editing”, and “knock-in allele” with orthopaedic-related terms: “musculoskeletal”, “bone”, “cartilage”, “tendon”, and “skeletal muscle”.
The origins of CRISPR-Cas9-mediated genome editing
CRISPR sequences present in the genomes of prokaryotes and their role as a defence mechanism against bacteriophage infection form the basis of CRISPR-mediated genome editing.5 The CRISPR system in Escherichia coli can be considered a form of adaptive immunity response where a record of invading bacteriophage is stored and weaponized to destroy future infecting bacteriophage particles.6 The mechanism is clever and sophisticated. Following bacteriophage infection, bacterial CRISPR-associated (Cas) proteins cleave the foreign DNA into small fragments and insert them into defined regions of the E. coli genome. With sequence information representing phage DNA, the bacteria now have a record of a prior phage exposure. Upon re-exposure to a new phage, a Cas nuclease is guided to a newly invading phage using the captured phage sequence. The foreign DNA is then cut by the Cas thereby incapacitating the invader. Many different Cas isoforms assembling distinct CRISPR systems have been described from a diverse range of bacterial and archaeal genomes and there are undoubtedly more to be discovered.
However, these clever bacterial defence mechanisms are not the whole story because bacteriophage have devised a way to fight back. As part of a type of molecular ‘arms race’, phage have developed a series of anti-CRISPR strategies.7,8 These can take several forms where some act to prevent the Cas molecule and wider complex from binding to DNA and others inhibit the Cas complex from assembling.9 Researchers have begun to utilize these molecules to further expand the utility of CRISPR systems by adding another layer of regulation.10-12
The basics of CRISPR-mediated genome editing in eukaryotic cells
While three major CRISPR systems have been described (types I, II, and III), it is the type II CRISPR/Cas system that has been the main system adapted for mammalian genome engineering.13 Each system employs the same basic mechanism of DNA editing but is constituted from a variety of proteins and isoforms.14 Jinek et al3 first demonstrated that CRISPR could be programmed for targeted DNA cleavage in vitro, in 2012. The following year, CRISPR-based genome editing in mammalian cells grown in culture was described.15,16 The enormous success and widespread use of CRISPR-based systems is reflected in the more than 19,000 PubMed articles listed since 2012. Despite this dominance of type II CRISPR systems, type I systems are actually more widespread in nature and are now being explored to expand the CRISPR system repertoire, and more innovations from this class are expected.
Successful gene editing relies on two different DNA repair pathways in eukaryotic cells: non-homologous end joining (NHEJ) and homology-directed repair (HDR) (Figure 1). Both repair pathways begin the same way with the CRISPR-Cas9 complex binding to its DNA target as specified by guide RNA (gRNA; green in Figure 1). The Cas9 nuclease binds to the RNA scaffold sequence and cuts DNA with nucleotide level specificity to create a double-strand break in the DNA sequence. The cellular DNA repair process then uses one of two mechanisms, NHEJ or HDR, to repair the double-strand break.
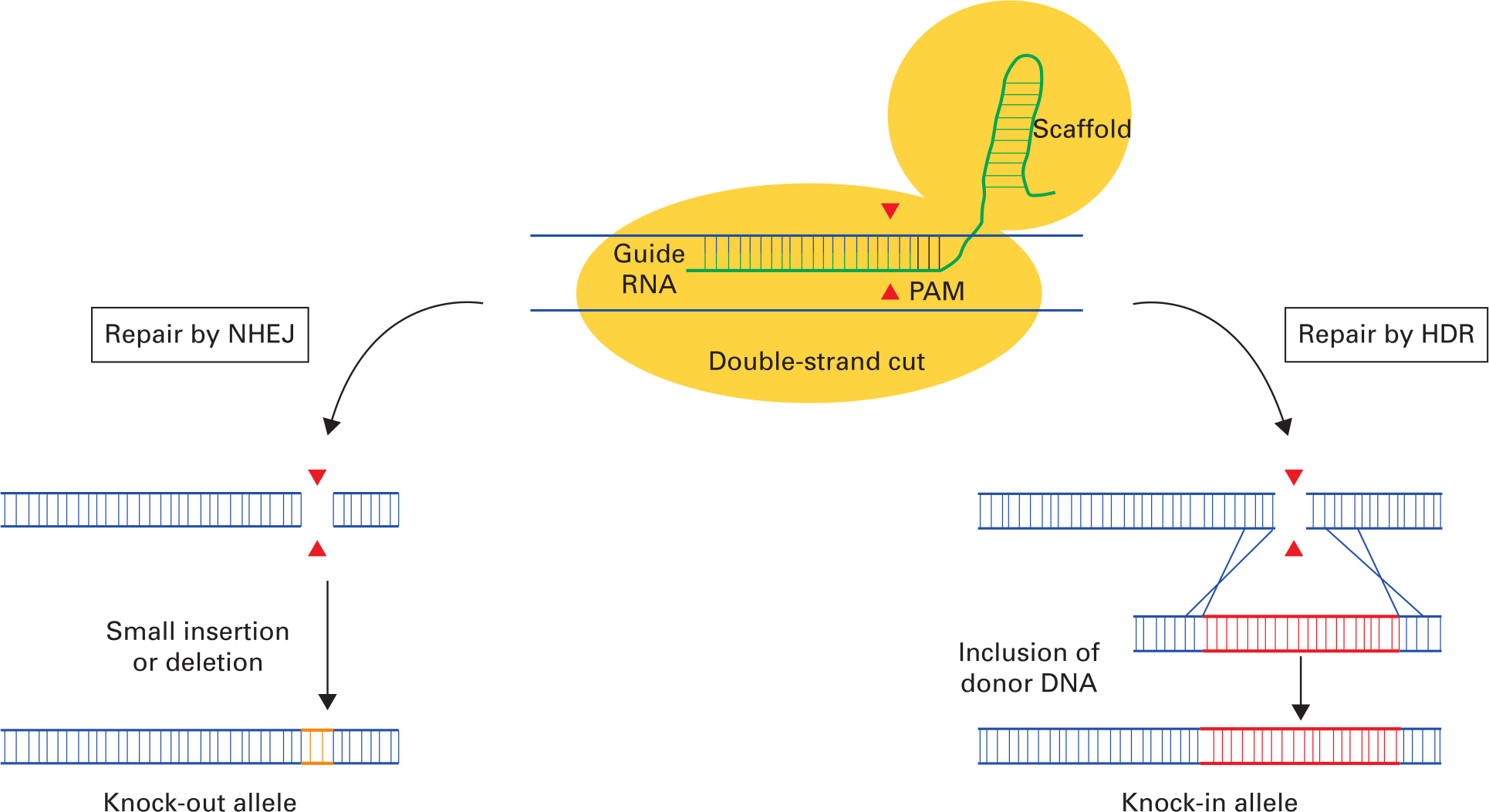
Fig. 1
Basic outline of Cas9 genome editing involving the two major DNA repair pathways: non-homologous end-joining (NHEJ) and homology-directed repair (HDR). The initial step is for the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 complex to bind to its DNA target. The CRISPR-Cas9 complex comprises Cas9 endonuclease (yellow) and single guide RNA (gRNA) and RNA scaffold (green) that binds to a specific DNA dictated by the gRNA. DNA cleavage will only occur when the gRNA is adjacent to the protospacer adjacent motif (PAM) recognition sequence. Following double-strand cleavage by the Cas9 endonuclease, DNA repair can be mediated by one of two possible mechanisms: NHEJ or HDR. Repair by NHEJ (left pathway) leads to the inclusion of DNA insertions or deletions (orange) resulting in a reading-frame shift, leading to a premature stop codon in the downstream sequence and the ‘knocking-out’ out of the targeted allele. Repair by HDR (right pathway) in the presence of donor DNA (red) targeted to the cut site results in insertion of the donor DNA sequence at the site of the DNA strand break and a ‘knock-in’ allele. Image adapted from Cribbs AP, Perera SMW. Science and Bioethics of CRISPR-Cas9 Gene Editing: An Analysis Towards Separating Facts and Fiction. Yale J Biol Med. 2017;90(4):625–634.
Non-homologous end joining
Following double-strand cleavage of DNA by Cas nucleases, the dominant repair process is NHEJ. However, NHEJ is relatively error-prone and DNA insertions and deletions (indels) in the target gene occur at the cut site. By adding or removing short sections of DNA, these indels often result in disruptions to the reading-frame of the protein coding region leading to a scrambled protein downstream from the altered sequence. Typically, a premature stop codon occurs downstream of the scrambled sequence and the mutant messenger RNA (mRNA) sequence is degraded via a process called nonsense-mediated mRNA degradation before the mutant mRNA can be translated into protein. This results in the so-called gene ‘knock-out’ (KO). Nonsense-mediated mRNA decay is a major evolutionary backup plan to minimize the consequences of aberrant protein production by recognizing the presence of premature stop codons.17
The three elements required for gene KO by NHEJ are gRNA sequence specific to target the region to be cut, RNA scaffold sequence for Cas9 binding, and Cas9 protein for DNA cleavage. In addition, a protospacer adjacent motif is required three to four nucleotides downstream from the region targeted by the gRNA. DNA cleavage will only occur when the gRNA is adjacent to a protospacer adjacent motif recognition sequence. Note that different Cas isoforms can have different protospacer adjacent motif recognition sequences.
These elements can be provided in one or two plasmid vectors or supplied as a ribonucleoprotein (RNP) complex. With the plasmid approach, the gRNA and scaffold are cloned into a plasmid DNA background together with the Cas9 gene under control of a viral promotor that directs robust gene expression in mammalian cells. The plasmid(s) are transfected into target cells and the inclusion of a selectable marker in the plasmid allows the possibility of clonally selecting transfected cells. With the ribonucleoprotein approach, the gRNA and scaffold sequence is prepared as RNA and mixed with Cas9 protein to create a RNA-protein hybrid. This complex is then transfected into cells. Since it is unknown how efficiently individual gRNA bind to target sequences, it is common to include several different gRNAs against the same target gene to maximize editing efficiency. For clinical applications, the adeno-associated virus is a Food and Drug Administration (FDA)-approved gene delivery vehicle where all the CRISPR elements are packed into viral particles and used to transduce target mammalian cells.18
Homology-directed repair
The HDR process allows the introduction of an endogenous DNA sequence at the site of the Cas9-mediated DNA double-strand break. If a segment of donor DNA is provided at the time of DNA cleavage, the host cell incorporates the introduced DNA into the cut site to create a ‘knock-in’ (KI) allele. The presence of homology arms designed to anneal to at least 1 kB of sequence either side of the cut site ensures DNA recombination and efficient homology-directed repair. The ability to KI exogenous DNA sequence is a particularly exciting advance because there is an endless variety of modifications that can be introduced including reporter sequences, specific mutations, or modified sequences (such as loxP/FRT sites) for tissue or temporal control of gene deletion. While the efficiency of genome modification by HDR is much less than that of NHEJ, HDR allows the creation of a variety of modifications with unprecedented speed and specificity than was previously possible.
Recent advances in CRISPR systems
The design of CRISPR genome editing systems is straightforward and within the reach of most molecular biology laboratories with many of the reagents available commercially. Basic laboratory equipment, such as polymerase chain reaction (PCR) machines, DNA gel running and visualization equipment, and tissue culture facilities, is required. Advances occur on a regular basis that increase the functionality of CRISPR through the engineering of specific elements of CRISPR systems to the discovery of new CRISPR systems in different prokaryotic species. Some of the key improvements are shown in Figure 2 and described in the following section.
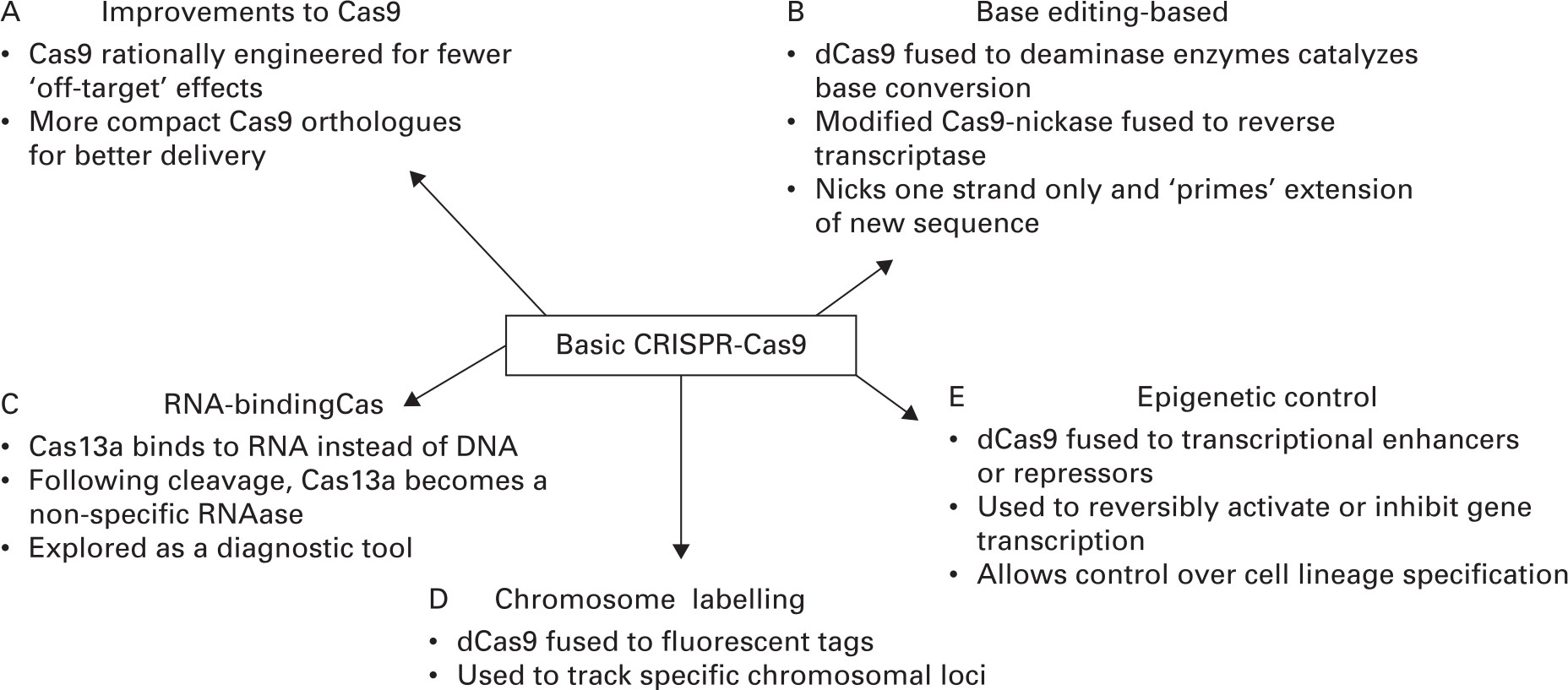
Fig. 2
Recent advances in clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9. The CRISPR-Cas9 system and its ability to target specific genomic loci has been adapted for a range of new applications. These include improvements to the Cas9 nuclease itself (A), active or inactive Cas9 fused to different protein modules for enhanced functionality (B, D, and E), and Cas isoforms that have high affinity for RNA instead of DNA (C). These advances have been applied to all the musculoskeletal tissues including cartilage, bone, and muscle.
High fidelity Cas9 isoforms
The incidence of unintended edits is a major concern for CRISPR-Cas9 genome editing.15,19 This refers to the rate of off-target edits due to the gRNA binding to closely related sequences in the genome besides the intended target. Estimations of off-target effects vary but appear to be dependent on individual gRNAs and the Cas nuclease isoform used. This area is under intense investigation. Since the intention is to use CRISPR for human therapies, safety is a paramount consideration. The original spCas9(1.1) (Streptococcus pyogenes Cas9) nuclease used in genome editing was found to have detectable unintended edits.15,20 Attempts have been made in recent years to rationally design better Cas isoforms by examining the 3D structure of Cas and defining the molecular points of contact it makes with DNA targets. These have led to a proliferation of spCas9 variants with very low off-target effects including enhanced-Cas9,21 HF1-Cas9,22 and HiFi-Cas9.23
Compact Cas9 orthologues
Multiple Cas9 orthologues have been identified with different properties compared to the spCas9 workhorse. Many of these are smaller than spCas9 and more easily packaged into viral vectors such as adeno-associated virus, which have limited cargo-carrying capacity.18 For example, Cas9 from Staphylococcus aureus (SaCas9) has similar editing efficiencies to spCas9 but is 350 amino acids smaller and therefore more easily packaged into delivery vectors.24 Similarly, CasX and CasY25,26 are smaller than spCas9. In addition, since they were isolated from archaeal bacterial species not normally pathogenic to humans, there should be no adaptive immune response that may be an issue for other Cas9 isoforms isolated from S. aureus, for example.27
Several Cas9 molecules have been identified that utilize different protospacer adjacent motif recognition sequences. For example, Cpf1 (Cas12a) is found in a widespread array of bacterial species and has a different (T-rich) protospacer adjacent motif recognition sequence compared to the (G-rich) Cas9 protospacer adjacent motif sequence.28 In addition, Cpf1 cuts in a staggered pattern allowing the possibility of directional gene transfer via HDR. Off-target effects are also reported to be lower in Cpf1 compared to spCas9.29,30
Base editing using CRISPR
Several CRISPR systems have been identified that do not use NHEJ or HDR but instead change individual bases without cutting the nucleic acid strand.31 The basis of this approach is the use of dead or catalytically inactive Cas9 (dCas9) fused to a cytidine deaminase enzyme that retains the ability to be reprogrammed with a gRNA. The fusion does not cut DNA but instead mediates the conversion of cytidine to uridine, thereby creating a C to G, or G to A base substitution. Several new refinements to the base editing approach have been developed that expand usefulness with fewer off-target effects,32 expanded protospacer adjacent motif specificities,33 and the development of more efficient editing variants.34,35
Prime CRISPR-Cas9
Late in 2019, a novel approach was described called ‘prime editing’, which promises to have fewer shortcomings and correct a large proportion of known pathogenic variants in humans.36 Prime editing, derived from base editing described above, differs from conventional CRISPR editing in two ways. A modified Cas9 nickase fused to reverse transcriptase cuts only one strand of the double-stranded target sequence instead of cutting both strands as conventional Cas9 does. Then a gRNA, which contains the sequence to be introduced, is added to the genome at the nicked site and inserted using the reverse transcriptase module attached to the modified Cas9 protein. Importantly, since only one strand is cut the technique promises to have fewer off-target effects. In general, prime editing is more precise and versatile than base editing. The extent to which prime editing will be useful remains to be determined but this is an exciting development in a field littered with significant discoveries.
RNA-binding Cas isoforms
CRISPR is a system that can be programmed to target specific stretches of the genome and to edit DNA via the Cas9 endonuclease at precise locations. Within the past two years, several RNA-binding Cas proteins have been identified that function to cleave single-stranded RNA molecules such as mRNA, rather than DNA. Cas13a (also named c2c2)37 utilizes the same gRNA system employed by Cas9-mediated editing but the RNA-Cas isoforms share no protein homology with DNA-Cas versions. Interestingly, unlike Cas9 systems where following cleavage the enzyme reverts to an inactive state, Cas13a retains enzymatic activity and becomes a non-specific ribonuclease.37 RNA-binding Cas riboendonucleases have expanded the functionality of CRISPR systems and have been used for RNA knockdown applications and adapted for pathogen detection.37,38
CRISPR-mediated epigenetic control
The targeting capacity of CRISPR system has been used to activate and repress gene transcription. The potential to reprogramme cell lineage specification via controlling gene transcription has wide-reaching applications for regenerative medicine, disease modelling, and even drug screening to name a few. The central idea is that dead-Cas9, where the catalytic domain that cuts DNA is inactivated, is fused with transcription activator or repressor domains. This dCas9 fusion is targeted to promotor regions to enhance or inhibit transcription in a reversible manner.
Much of the pioneering work has been conducted in the Gersbach laboratory at Duke University to control developmental events. This was first demonstrated in 2014 when Chakraborty et al39 showed that targeted activation of the myogenic transcription factor, MyoD, in mouse embryonic fibroblasts using gene activation domains fused to Cas9 reprogrammed the cells to differentiate into skeletal myocytes. More recent innovations involve repurposing type I CRISPR systems for programmable gene regulation.40
Chromosomal tracking
CRISPR can be used to dynamically track specific genomic loci. As with the examples described above dCas9 is used but is fused to fluorescent tags. This is similar to fluorescent in situ hybridization but with several advantages: probes are easy to make, and labelling can be done in live cells. The gRNA and Cas9 will bind to specific regions. Using dead-Cas9 fused to a fluorescent marker like GFP, researchers have turned dCas9 into a customizable DNA labeler compatible with fluorescence microscopy in living cells. CRISPR imaging has numerous advantages over other imaging techniques, including ease of implementation due to the simplicity of gRNA design, programmability for different genomic loci, capability of detecting multiple genomic loci, and compatibility with live cell imaging.
CRISPR applications in basic orthopaedic research
With this broad array of CRISPR systems that are available from gene KO and KI modifications to base editing and epigenetic modifications, what impact have these tools had on research into the musculoskeletal tissues including bone, cartilage, and skeletal muscle?
Knock-out editing in vitro
Since it is relatively straightforward to KO specific genes in cell lines using the NHEJ mechanism (Figure 1) and within the capabilities of most orthopaedic molecular biology laboratories, most work in the bone and joint field has been directed towards editing cell lines. Several examples of this are described below.
CRISPR editing has been particularly useful for investigating the role of specific genes in osteosarcoma cells’ progression in humans cells including cell cycle genes such as CDK11,41 factors involved in collagen biosynthesis such as GLT25D1,42 metastasis suppressor genes in mouse cells such as Srgap2,43 and drug resistance.44 With the availability of ‘libraries’ of gene KO reagents, it is now possible to conduct large-scale cellular screening for novel activators/suppressors of osteosarcoma. This is a fruitful area of research that should yield new and exciting insights into osteosarcoma biology.
For the study of events critical for osteoclastogenesis, mouse RAW264 cells deleted for Zscan10 and differentiated into osteoclasts by RANKL stimulation were found to have increased osteoclast activity.45 Potential therapeutic factors were then tested for ability to reduce osteoclastogenesis for the treatment of low bone mineral density.
The role of specific genes in chondrocyte plasticity and regeneration has been explored. For example, deletion of the gap junction channel protein connexin43 attenuates cellular senescence and promotes chondrocyte regeneration in osteoarthritis.46 The power of CRISPR-mediated gene deletion to ‘cleanly’ dissect the contribution of closely related family members to cartilage biology was nicely demonstrated in a study of the WNT signalling pathway members.47 Here, CRISPR was used to delete three WNT family members independently, thereby allowing the investigators to separately determine the contribution of each WNT factor to chondrocyte differentiation.47
Knock-out editing in animals
The natural follow-up from in vitro studies is to confirm findings and recapitulate human genetic variants in rodent models. Gene KO alleles generated by CRISPR are quicker and easier to make than ‘classic’ transgenic technologies resulting in notable cost and time savings to investigators with F1 animals at the screening stage in as little as three months, including construct generation time.
In the area of orthopaedic cancer, the functions of key genes involved in osteosarcoma progression such as Tp5348 and Era49 have been explored in mouse models using CRISPR-mediated gene KOs, and similar KO experiments in rats have also been described.50
Correction of disease-causing mutations in developing musculoskeletal tissues is a major research goal. Miao et al51 corrected the Fgfr3-glycine-to-arginine mutation that causes chondrodysplasia in mice and optimized correction by tweaking their system to maximize the efficiency of correction. CRISPR-mediated KO of the anoctamin-5 gene resulted in a phenotype that resembles the human disease gnathodiaphyseal dysplasia, a rare skeletal disorder that is characterized by mandibular lesions, bone fragility, and sclerosis of tubular bones.52
Many genes function in multiple tissues and physiological processes. Conditional KO mice are a useful tool to isolate KO effects in one tissue from effects in other tissues. Wee et al53 generated neuropeptide Y (NPY)-floxed (NPYflox) mice using CRISPR technology, allowing them to study global neuropeptide Y-KO mice alongside tissue-specific neuropeptide Y-KO mice, thereby providing a versatile tool for the study of neuropeptide Y in skeletal development.
Other animal models besides mice and rats are being used in orthopaedic research and to provide critical supporting data initially generated in human studies. Although lacking a weight-bearing skeleton, zebrafish have been used to investigate skeletal development for several decades, and CRISPR-mediated gene editing has been useful for the rapid generation of mutant variants for modelling human disease and the analysis of skeletal development. For example, whole-exome sequencing and linkage analysis identified three disease-causing missense variants in the MAPK7 gene human populations with adolescent idiopathic scoliosis.54 CRISPR-mediated deletion of mapk7 in zebrafish recapitulated the characteristics of idiopathic scoliosis, thereby confirming the findings in humans. Similarly, variants in the ATP6V1H gene are associated with short stature and osteoporosis in humans. Loss of function mutants in the zebrafish atp6v1h gene recapitulated the bone phenotype. The phenotype was recovered with inhibitors of specific matrix metalloproteinases, suggesting a new set of therapeutic targets for this disorder.55 Epigenetic regulators such as histone deacetylases and acetyltransferases and their role in skeletal development have also been investigated using CRISPR in zebrafish.56
The majority of genome manipulations have been conducted in small rodents, which lack features present in larger animal models such as Haversian canals and have different growth plate characteristics.57,58 CRISPR genome manipulation technologies now allow the exciting possibilities of answering orthopaedic-related questions in larger animal models that more closely resemble the human skeletal system in anatomy, cellular physiology, and the biomechanical environment. For example, deletion of DMP1 in rabbits is leading to a new understanding of hypophosphatemic rickets Also in rabbits, deletion of the PHEX gene, encoding a phosphate-regulating endopeptidase responsible for X-linked hypophosphatemia, recapitulates the features of rickets in humans and will be a good model of this disorder moving forward.59 The identification of a rabbit ROSA26 locus will further spur orthopaedic research in rabbits because of its use as a safe genomic harbor suitable for nuclease-mediated gene targeting.60
Gene knock-ins in vitro
KI models generated by the HDR pathway are more complicated to generate than KO models. However, compared to gene KOs, KIs provide additional functionality such as the inclusion of protein reporter sequences and the modelling of human mutations and potential variants (Figure 1). KIs utilize the HDR pathway provided a segment of donor DNA is available for insertion into the cut site. Typically, two DNA constructs are required, one to carry the gRNA and Cas sequences necessary for DNA cleavage and a second that contains the endogenous sequence to be inserted at the cut site. Due to this added complexity, the generation of KI models is less efficient than that of KO models and requires more screening for the desired editing.
Bone marrow-resident mesenchymal stem cells are the starting point for many tissue engineering strategies because they can be induced to differentiate into bone, cartilage, and adipose tissue. To generate an immortalized mesenchymal stem cell line, Hu et al61 knocked-in the SV40 sequence into the Rosa26 locus using CRISPR. The resulting cells are immortalized and can differentiate into multiple cell types. Importantly, the immortalization is reversible via removal of the SV40 sequence by flip recombinase (FLP)-mediated excision. Generating a supply of reversibly immortalized mesenchymal stem cells represents a powerful technical advance that will have a host of tissue engineering and regeneration applications.
Novel tools are being developed using CRISPR-based KI approaches. For example, human-induced pluripotent stem cells are being explored as a starting point in tissue regeneration strategies. A COL2A1-driven green fluorescent protein (GFP) reporter was knocked-in to a human-induced pluripotent stem cell line using CRISPR. This allowed the identification and purification of chondrogenic precursor cells from mixed populations of stem cells with potential applications for tissue engineering and in drug screens for factors that promote cartilage production.62
There have been several reports of KI mutations in mice.63,64 Gain-of-function truncation mutations in NOTCH3 in humans result in lateral meningocele syndrome, a rare disorder characterized by neurological complications and osteoporosis. A stop codon cassette was knocked-in in mice at the same location as a human mutation and the resulting mice developed cancellous and cortical bone osteopenia without neurological complications.63
Patients with the MAB21L2 (arginine to cysteine at amino acid 51) mutation have eye abnormalities, skeletal dysplasia, and intellectual disability. To understand the pathology further, the R51C mutation was introduced into mice by CRISPR. The Mab21l2R51C/+ mice have eyeless phenotype and skeletal abnormalities including a joint fusion phenotype where the humeri are fused with the radii, and the femur is fused with the tibia. These features model the human eye and bone phenotypes in humans.64
Knock-in large animal models
The availability of high-quality whole genome sequences for major mammalian species coupled with advances in reproductive biology and embryology means that larger, more orthopaedically relevant species can now be edited using CRISPR. The first large animal study using a KI CRISPR approach was accomplished in sheep in 2018. Williams et al65 developed a model of hypophosphatasia by knocking-in a known disease-causing human mutation, resulting in an amino acid substitution at position 359, into the sheep alkaline phosphatase-like gene. The resulting sheep have mineralization defects consistent with human hypophosphatasia. This landmark experiment establishes sheep as a model for studying skeletal development and maintenance, and paves the way for other domesticated species to be manipulated given an adequate understanding of the relevant reproductive biology.
Epigenetic regulation
Another approach targeted epigenetic marks to create a genetic switch that allows for programmable and reversible gene activation in virtually any gene. In one early ‘proof-of-concept’ study, dCas9 was fused to the catalytic core of the human acetyltransferase p300 domain.66 The fusion protein catalyzes acetylation of histone H3 lysine 27 at its target sites. This leads to gene activation at both distal and proximal promoters. The investigators successfully enhanced transcription of key developmental transcription factors at both promotor sites and more distal enhancer sites. Unlike targeting epigenetic marks with drugs or inhibitors of histone deacetylases or DNA methyltransferase, this approach will be useful for targeting individual loci. Mochizuki et al67 developed a combinatorial CRISPR-Cas9 approach for the identification of cartilage-specific Sox9 distal enhancers important for skeletal development. Using a combination of dCas9-mediated epigenetic silencing and the generation of enhancer deletion mice, chromatin immunoprecipitation, and mass spectrometry, the authors identified a far-upstream cis-element that regulates cartilage-specific Sox9 expression and subsequent skeletal development.
As these examples demonstrate, the CRISPR toolbox is diverse and continually expanding, allowing a large range of genome manipulations. How are these tools impacting orthopaedic clinical research?
Perspectives on CRISPR and orthopaedic clinical practice
The use of CRISPR as a tool for genome modification is transforming orthopaedic research by accelerating the pace of research, particularly for modelling disease-causing mutations in vitro and in vivo. While there are significant problems to overcome such as off-target effects and lack of effective delivery strategies, the field moves ahead with blinding speed. Despite these impressive developments in cell and animal models, widespread clinical use is some time off and will likely be approved on a case-by-case basis with the need for each therapy to be validated in cells, animals, and later in humans for safety.
Provided off-target editing effects can be greatly minimized or eliminated, the delivery of gene therapies to the intended cells remains a stubborn technical problem and is the rate-limiting step in the development of new treatments. This is particularly true for musculoskeletal tissues where access is difficult with some cell types, such as chondrocytes, being refractory to transfection. At the clinical level, the experience with targeting of gene therapies to musculoskeletal tissues has generally been disappointing. For example, the delivery of antisense oligonucleotide-based drugs to skeletal muscle for Duchenne muscular dystrophy has largely been unsuccessful in clinical trials.68 It is noteworthy that the FDA-approved CRISPR-based treatment targets retinal tissue, which is easily accessible, and reagents can be deposited directly onto the relevant eye structures. With orthopaedic tissues, the extracellular matrix also presents a significant barrier to delivery. The field must wait for safe and effective reagent delivery methods. Most popular approaches, including for musculoskeletal applications,69 utilize adeno-associated viral and lentiviral-based methods because these viruses have evolved machinery to efficiently infect mammalian cells. Adeno-associated virus has been approved by the FDA for human use, and most CRISPR-based clinical applications will use adeno-associated virus as the delivery vector in the foreseeable future. The drawbacks with viral vectors include packaging capacity, immune response, and, for lentivirus, lack of control over integration sites. This lack of cargo-carrying capacity may be an issue for KI applications where large donor sequences to be knocked-in are required and other vector systems that allow for the packaging of larger cargoes need to be explored.
In the orthopaedic realm, the areas most likely to have an impact on clinical practice are in drug discovery and the development of large animal models. The ability to make a single, discrete change in the genome of cells in vitro, or model organisms in vivo, lends itself to drug discovery. The availability of libraries containing CRISPR-generated mutants makes it possible to conduct large-scale genome-wide screens using phenotypic readouts at the cellular level.70 CRISPR screening will be a powerful tool for drug discovery.
With the goal of accurately replicating human orthopaedic pathologies, large animal models are critically important. The first demonstration of an orthopaedic disease model in sheep is significant because sheep and other large animals are anatomically and biomechanically more relevant for answering orthopaedic research questions than rodents. While a transgenic sheep core will need significant resources to establish and maintain, one can envisage that a rich trove of findings relevant to human orthopaedic conditions would result from such a resource.
In conclusion, while the future looks good for basic orthopaedic research with CRISPR-based generation of models from zebrafish to mouse and recently, rabbits and sheep, at the clinical level no CRISPR-based therapies exist for orthopaedic conditions. Here, the rate-limiting step is the lack of efficient delivery options to the relevant cells and tissues, such as bone and cartilage, which have specialized extracellular matrices that are a significant barrier to delivery. These delivery limitations will need to be overcome if effective CRISPR-based therapies for orthopaedic disorders are to be implemented. The light at the end of the tunnel is that new delivery strategies are under intense development ranging from nanoparticles,71 nucleopeptides,72 and polymers73 to new viral vectors.74 This a rapidly moving field. Stay tuned…
References
1. Miller JC , Tan S , Qiao G , et al. A TALE nuclease architecture for efficient genome editing . Nat Biotechnol . 2011 ; 29 ( 2 ): 143 – 148 . Crossref PubMed Google Scholar
2. Zhang F , Cong L , Lodato S , et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription . Nat Biotechnol . 2011 ; 29 ( 2 ): 149 – 153 . Crossref PubMed Google Scholar
3. Jinek M , Chylinski K , Fonfara I , et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity . Science . 2012 ; 337 ( 6096 ): 816 – 821 . Crossref PubMed Google Scholar
4. Ran FA , Hsu PD , Wright J , et al. Genome engineering using the CRISPR-Cas9 system . Nat Protoc . 2013 ; 8 ( 11 ): 2281 – 2308 . Google Scholar
5. Sontheimer EJ , Barrangou R . The Bacterial Origins of the CRISPR Genome-Editing Revolution . Hum Gene Ther . 2015 ; 26 ( 7 ): 413 – 424 . Crossref PubMed Google Scholar
6. Barrangou R , Fremaux C , Deveau H , et al. CRISPR provides acquired resistance against viruses in prokaryotes . Science . 2007 ; 315 ( 5819 ): 1709 – 1712 . Crossref PubMed Google Scholar
7. Bondy-Denomy J , Pawluk A , Maxwell KL , Davidson AR . Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system . Nature . 2013 ; 493 ( 7432 ): 429 – 432 . Crossref PubMed Google Scholar
8. Hwang S , Maxwell KL . Meet the Anti-CRISPRs: Widespread Protein Inhibitors of CRISPR-Cas Systems . CRISPR J . 2019 ; 2 ( 1 ): 23 – 30 . Crossref PubMed Google Scholar
9. Bondy-Denomy J , Garcia B , Strum S , et al. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins . Nature . 2015 ; 526 ( 7571 ): 136 – 139 . Crossref PubMed Google Scholar
10. Lee J , Mou H , Ibraheim R , et al. Tissue-restricted genome editing in vivo specified by microRNA-repressible anti-CRISPR proteins . RNA . 2019 ; 25 ( 11 ): 1421 – 1431 . Crossref PubMed Google Scholar
11. Nakamura M , Srinivasan P , Chavez M , et al. Anti-CRISPR-mediated control of gene editing and synthetic circuits in eukaryotic cells . Nat Commun . 2019 ; 10 ( 1 ): 194 . Crossref PubMed Google Scholar
12. Bubeck F , Hoffmann MD , Harteveld Z , et al. Engineered anti-CRISPR proteins for optogenetic control of CRISPR-Cas9 . Nat Methods . 2018 ; 15 ( 11 ): 924 – 927 . Crossref PubMed Google Scholar
13. Makarova KS , Wolf YI , Alkhnbashi OS , et al. An updated evolutionary classification of CRISPR-Cas systems . Nat Rev Microbiol . 2015 ; 13 ( 11 ): 722 – 736 . Crossref PubMed Google Scholar
14. Makarova KS , Haft DH , Barrangou R , et al. Evolution and classification of the CRISPR-Cas systems . Nat Rev Microbiol . 2011 ; 9 ( 6 ): 467 – 477 . Crossref PubMed Google Scholar
15. Cong L , Ran FA , Cox D , et al. Multiplex genome engineering using CRISPR/Cas systems . Science . 2013 ; 339 ( 6121 ): 819 – 823 . Crossref PubMed Google Scholar
16. Mali P , Yang L , Esvelt KM , et al. RNA-guided human genome engineering via Cas9 . Science . 2013 ; 339 ( 6121 ): 823 – 826 . Crossref PubMed Google Scholar
17. Kurosaki T , Popp MW , Maquat LE . Quality and quantity control of gene expression by nonsense-mediated mRNA decay . Nat Rev Mol Cell Biol . 2019 ; 20 ( 7 ): 406 – 420 . Crossref PubMed Google Scholar
18. Wang D , Tai PWL , Gao G . Adeno-associated virus vector as a platform for gene therapy delivery . Nat Rev Drug Discov . 2019 ; 18 ( 5 ): 358 – 378 . Crossref PubMed Google Scholar
19. Hendel A , Fine EJ , Bao G , Porteus MH . Quantifying on- and off-target genome editing . Trends Biotechnol . 2015 ; 33 ( 2 ): 132 – 140 . Crossref PubMed Google Scholar
20. Fu Y , Foden JA , Khayter C , et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells . Nat Biotechnol . 2013 ; 31 ( 9 ): 822 – 826 . Crossref PubMed Google Scholar
21. Slaymaker IM , Gao L , Zetsche B , et al. Rationally engineered Cas9 nucleases with improved specificity . Science . 2016 ; 351 ( 6268 ): 84 – 88 . Crossref PubMed Google Scholar
22. Kleinstiver BP , Pattanayak V , Prew MS , et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects . Nature . 2016 ; 529 ( 7587 ): 490 – 495 . Crossref PubMed Google Scholar
23. Vakulskas CA , Dever DP , Rettig GR , et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells . Nat Med . 2018 ; 24 ( 8 ): 1216 – 1224 . Crossref PubMed Google Scholar
24. Ran FA , Cong L , Yan WX , et al. In vivo genome editing using Staphylococcus aureus Cas9 . Nature . 2015 ; 520 ( 7546 ): 186 – 191 . Crossref PubMed Google Scholar
25. Burstein D , Harrington LB , Strutt SC , et al. New CRISPR-Cas systems from uncultivated microbes . Nature . 2017 ; 542 ( 7640 ): 237 – 241 . Crossref PubMed Google Scholar
26. Liu JJ , Orlova N , Oakes BL , et al. CasX enzymes comprise a distinct family of RNA-guided genome editors . Nature . 2019 ; 566 ( 7743 ): 218 – 223 . Crossref PubMed Google Scholar
27. Charlesworth CT , Deshpande PS , Dever DP , et al. Identification of pre-existing adaptive immunity to Cas9 proteins in humans . Nat Med . 2019 ; 25 ( 2 ): 249 – 254 . Google Scholar
28. Zetsche B , Gootenberg JS , Abudayyeh OO , et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system . Cell . 2015 ; 163 ( 3 ): 759 – 771 . Crossref PubMed Google Scholar
29. Kim D , Kim J , Hur JK , et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells . Nat Biotechnol . 2016 ; 34 ( 8 ): 863 – 868 . Crossref PubMed Google Scholar
30. Kleinstiver BP , Tsai SQ , Prew MS , et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells . Nat Biotechnol . 2016 ; 34 ( 8 ): 869 – 874 . Crossref PubMed Google Scholar
31. Komor AC , Kim YB , Packer MS , Zuris JA , Liu DR . Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage . Nature . 2016 ; 533 ( 7603 ): 420 – 424 . Crossref PubMed Google Scholar
32. Rees HA , Komor AC , Yeh WH , et al. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery . Nat Commun . 2017 ; 8 : 15790 . Crossref PubMed Google Scholar
33. Kim YB , Komor AC , Levy JM , et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions . Nat Biotechnol . 2017 ; 35 ( 4 ): 371 – 376 . Crossref PubMed Google Scholar
34. Nishida K , Arazoe T , Yachie N , et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems . Science . 2016 ; 353 ( 6305 ): 353 . Crossref PubMed Google Scholar
35. Komor AC , Zhao KT , Packer MS , et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity . Sci Adv . 2017 ; 3 ( 8 ): eaao4774 . Crossref PubMed Google Scholar
36. Anzalone AV , Randolph PB , Davis JR , et al. Search-and-replace genome editing without double-strand breaks or donor DNA . Nature . 2019 ; 576 ( 7785 ): 149 – 157 . Google Scholar
37. Abudayyeh OO , Gootenberg JS , Essletzbichler P , et al. RNA targeting with CRISPR-Cas13 . Nature . 2017 ; 550 ( 7675 ): 280 – 284 . Crossref PubMed Google Scholar
38. Gootenberg JS , Abudayyeh OO , Lee JW , et al. Nucleic acid detection with CRISPR-Cas13a/C2c2 . Science . 2017 ; 356 ( 6336 ): 438 – 442 . Crossref PubMed Google Scholar
39. Chakraborty S , Ji H , Kabadi AM , et al. A CRISPR/Cas9-based system for reprogramming cell lineage specification . Stem Cell Reports . 2014 ; 3 ( 6 ): 940 – 947 . Crossref PubMed Google Scholar
40. Pickar-Oliver A , Black JB , Lewis MM , et al. Targeted transcriptional modulation with type I CRISPR-Cas systems in human cells . Nat Biotechnol . 2019 ; 37 ( 12 ): 1493 – 1501 . Crossref PubMed Google Scholar
41. Feng Y , Sassi S , Shen JK , et al. Targeting CDK11 in osteosarcoma cells using the CRISPR-Cas9 system . J Orthop Res . 2015 ; 33 ( 2 ): 199 – 207 . Crossref PubMed Google Scholar
42. Baumann S , Hennet T . Collagen Accumulation in Osteosarcoma Cells lacking GLT25D1 Collagen Galactosyltransferase . J Biol Chem . 2016 ; 291 ( 35 ): 18514 – 18524 . Crossref PubMed Google Scholar
43. Marko TA , Shamsan GA , Edwards EN , et al. Slit-Robo GTPase-Activating Protein 2 as a metastasis suppressor in osteosarcoma . Sci Rep . 2016 ; 6 : 39059 . Crossref PubMed Google Scholar
44. Xiao Z , Wan J , Nur AA , et al. Targeting CD44 by CRISPR-Cas9 in Multi-Drug Resistant Osteosarcoma Cells . Cell Physiol Biochem . 2018 ; 51 ( 4 ): 1879 – 1893 . Crossref PubMed Google Scholar
45. Yanagihara Y , Inoue K , Saeki N , et al. Zscan10 suppresses osteoclast differentiation by regulating expression of Haptoglobin . Bone . 2019 ; 122 : 93 – 100 . Crossref PubMed Google Scholar
46. Varela-Eirín M , Varela-Vázquez A , Guitián-Caamaño A , et al. Targeting of chondrocyte plasticity via connexin43 modulation attenuates cellular senescence and fosters a pro-regenerative environment in osteoarthritis . Cell Death Dis . 2018 ; 9 ( 12 ): 1166 . Crossref PubMed Google Scholar
47. Pizzute T , He F , Zhang XB , Pei M . Impact of Wnt signals on human intervertebral disc cell regeneration . J Orthop Res . 2018 ; 36 ( 12 ): 3196 – 3207 . Crossref PubMed Google Scholar
48. Tang F , Min L , Seebacher NA , et al. Targeting mutant TP53 as a potential therapeutic strategy for the treatment of osteosarcoma . J Orthop Res . 2019 ; 37 ( 3 ): 789 – 798 . Crossref PubMed Google Scholar
49. Lillo Osuna MA , Garcia-Lopez J , El Ayachi I , et al. Activation of Estrogen Receptor Alpha by Decitabine Inhibits Osteosarcoma Growth and Metastasis . Cancer Res . 2019 ; 79 ( 6 ): 1054 – 1068 . Crossref PubMed Google Scholar
50. Suzuki H , Ito Y , Shinohara M , et al. Gene targeting of the transcription factor Mohawk in rats causes heterotopic ossification of Achilles tendon via failed tenogenesis . Proc Natl Acad Sci U S A . 2016 ; 113 ( 28 ): 7840 – 7845 . Crossref PubMed Google Scholar
51. Miao K , Zhang X , Su SM , et al. Optimizing CRISPR/Cas9 technology for precise correction of the Fgfr3-G374R mutation in achondroplasia in mice . J Biol Chem . 2019 ; 294 ( 4 ): 1142 – 1151 . Crossref PubMed Google Scholar
52. Wang X , Liu X , Dong R , et al. Genetic Disruption of Anoctamin 5 in Mice Replicates Human Gnathodiaphyseal Dysplasia (GDD) . Calcif Tissue Int . 2019 ; 104 ( 6 ): 679 – 689 . Crossref PubMed Google Scholar
53. Wee NKY , Sinder BP , Novak S , et al. Skeletal phenotype of the neuropeptide Y knockout mouse . Neuropeptides . 2019 ; 73 : 78 – 88 . Crossref PubMed Google Scholar
54. Gao W , Chen C , Zhou T , et al. Rare coding variants in MAPK7 predispose to adolescent idiopathic scoliosis . Hum Mutat . 2017 ; 38 ( 11 ): 1500 – 1510 . Crossref PubMed Google Scholar
55. Zhang Y , Huang H , Zhao G , et al. ATP6V1H Deficiency Impairs Bone Development through Activation of MMP9 and MMP13 . PLoS Genet . 2017 ; 13 ( 2 ): e1006481 . Crossref PubMed Google Scholar
56. DeLaurier A , Alvarez CL , Wiggins KJ . hdac4 mediates perichondral ossification and pharyngeal skeleton development in the zebrafish . PeerJ . 2019 ; 7 : e6167 . Crossref PubMed Google Scholar
57. Lelovas PP , Xanthos TT , Thoma SE , Lyritis GP , Dontas IA . The laboratory rat as an animal model for osteoporosis research . Comp Med . 2008 ; 58 ( 5 ): 424 – 430 . PubMed Google Scholar
58. Duranova H , Martiniakova M , Omelka R , et al. Changes in compact bone microstructure of rats subchronically exposed to cadmium . Acta Vet Scand . 2014 ; 56 : 64 . Crossref PubMed Google Scholar
59. Sui T , Yuan L , Liu H , et al. CRISPR/Cas9-mediated mutation of PHEX in rabbit recapitulates human X-linked hypophosphatemia (XLH) . Hum Mol Genet . 2016 ; 25 ( 13 ): 2661 – 2671 . Crossref PubMed Google Scholar
60. Yang D , Song J , Zhang J , et al. Identification and characterization of rabbit ROSA26 for gene knock-in and stable reporter gene expression . Sci Rep . 2016 ; 6 : 25161 . Crossref PubMed Google Scholar
61. Hu X , Li L , Yu X , et al. CRISPR/Cas9-mediated reversibly immortalized mouse bone marrow stromal stem cells (BMSCs) retain multipotent features of mesenchymal stem cells (MSCs) . Oncotarget . 2017 ; 8 ( 67 ): 111847 – 111865 . Crossref PubMed Google Scholar
62. Adkar SS , Wu C-L , Willard VP , et al. Step-Wise Chondrogenesis of Human Induced Pluripotent Stem Cells and Purification Via a Reporter Allele Generated by CRISPR-Cas9 Genome Editing . Stem Cells . 2019 ; 37 ( 1 ): 65 – 76 . Crossref PubMed Google Scholar
63. Canalis E , Yu J , Schilling L , Yee SP , Zanotti S . The lateral meningocele syndrome mutation causes marked osteopenia in mice . J Biol Chem . 2018 ; 293 ( 36 ): 14165 – 14177 . Crossref PubMed Google Scholar
64. Tsang SW , Guo Y , Chan LH , Huang Y , Chow KL . Generation and characterization of pathogenic Mab21l2(R51C) mouse model . Genesis . 2018 ; 56 ( 11-12 ): e23261 . Crossref PubMed Google Scholar
65. Williams DK , Pinzón C , Huggins S , et al. Genetic engineering a large animal model of human hypophosphatasia in sheep . Sci Rep . 2018 ; 8 ( 1 ): 16945 . Crossref PubMed Google Scholar
66. Hilton IB , D’Ippolito AM , Vockley CM , et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers . Nat Biotechnol . 2015 ; 33 ( 5 ): 510 – 517 . Crossref PubMed Google Scholar
67. Mochizuki Y , Chiba T , Kataoka K , et al. Combinatorial CRISPR/Cas9 Approach to Elucidate a Far-Upstream Enhancer Complex for Tissue-Specific Sox9 Expression . Dev Cell . 2018 ; 46 ( 6 ): 794 – 806 . Crossref PubMed Google Scholar
68. Lin GA , Agboola F , Otuonya I , et al. Deflazacort, eteplirsen, and golodirsen for Duchenne Muscular Dystrophy: Effectiveness and value - Evidence report. Institute for Clinical and Economic Review . 2019 : 115 – 133 . Google Scholar
69. Evans CH . The vicissitudes of gene therapy . Bone Joint Res . 2019 ; 8 ( 10 ): 469 – 471 . Crossref PubMed Google Scholar
70. Yu JSL , Yusa K . Genome-wide CRISPR-Cas9 screening in mammalian cells . Methods . 2019 ; 164–165 : 29 – 35 . Crossref PubMed Google Scholar
71. Chen J , Guo Z , Tian H , Chen X . Production and clinical development of nanoparticles for gene delivery . Mol Ther Methods Clin Dev . 2016 ; 3 : 16023 . Crossref PubMed Google Scholar
72. Wang H , Feng Z , Xu B . Supramolecular Assemblies of Peptides or Nucleopeptides for Gene Delivery . Theranostics . 2019 ; 9 ( 11 ): 3213 – 3222 . Crossref PubMed Google Scholar
73. Sung YK , Kim SW . Recent advances in the development of gene delivery systems . Biomater Res . 2019 ; 23 : 8 . Crossref PubMed Google Scholar
74. Gonçalves GAR , Paiva RMA . Gene therapy: advances, challenges and perspectives . Einstein (Sao Paulo). 2017 : 15(3):369 – 375 . Google Scholar
Author contributions
J. Fitzgerald: Wrote the manuscript.
Funding statement
No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
ICMJE COI statement
None declared.
Acknowledgements
The work was supported in part by the Orphan Diseases Centre, grant #MDBR-19 to 107-CMD.
Ethical review statement
This study did not require ethical approval.
© 2020 Author(s) et al. This is an open-access article distributed under the terms of the Creative Commons Attribution Non-Commercial No Derivatives (CC BY-NC-ND 4.0) licence, which permits the copying and redistribution of the work only, and provided the original author and source are credited. See https://creativecommons.org/licenses/by-nc-nd/4.0/.









