Abstract
Aims
Circular RNA (circRNA) S-phase cyclin A-associated protein in the endoplasmic reticulum (ER) (circSCAPER, ID: hsa_circ_0104595) has been found to be highly expressed in osteoarthritis (OA) patients and has been associated with the severity of OA. Hence, the role and mechanisms underlying circSCAPER in OA were investigated in this study.
Methods
In vitro cultured human normal chondrocyte C28/I2 was exposed to interleukin (IL)-1β to mimic the microenvironment of OA. The expression of circSCAPER, microRNA (miR)-140-3p, and enhancer of zeste homolog 2 (EZH2) was detected using quantitative real-time polymerase chain reaction and Western blot assays. The extracellular matrix (ECM) degradation, proliferation, and apoptosis of chondrocytes were determined using Western blot, cell counting kit-8, and flow cytometry assays. Targeted relationships were predicted by bioinformatic analysis and verified using dual-luciferase reporter and RNA immunoprecipitation (RIP) assays. The levels of phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT) pathway-related protein were detected using Western blot assays.
Results
CircSCAPER was highly expressed in OA cartilage tissues and IL-1β-induced chondrocytes. Knockdown of circSCAPER reduced IL-1β-evoked ECM degradation, proliferation arrest, and apoptosis enhancement in chondrocytes. Mechanistically, circSCAPER directly bound to miR-140-3p, and miR-140-3p inhibition reversed the effects of circSCAPER knockdown on IL-1β-induced chondrocytes. miR-140-3p was verified to target EZH2, and overexpression of miR-140-3p protected chondrocytes against IL-1β-induced dysfunction via targeting EZH2. Additionally, we confirmed that circSCAPER could regulate EZH2 through sponging miR-140-3p, and the circSCAPER/miR-140-3p/EZH2 axis could activate the PI3K/AKT pathway.
Conclusion
CircSCAPER promoted IL-1β-evoked ECM degradation, proliferation arrest, and apoptosis enhancement in chondrocytes via regulating miR-140-3p/EZH2 axis, which gained a new insight into the pathogenesis of OA.
Cite this article: Bone Joint Res 2022;11(2):61–72.
Article focus
-
The aim of this study was to provide a comprehensive understanding of the effects of circSCAPER on chondrocytes affected by osteoarthritis (OA).
Key messages
-
In vitro cultured human normal chondrocyte C28/I2 was exposed to interleukin (IL)-1β to mimic the microenvironment of OA, and circSCAPER enhanced IL-1β-induced inhibition of proliferation and promotion of apoptosis in chondrocytes via the microRNA (miR)-140-3p/enhancer of zeste homolog 2 (EZH2) axis.
Strengths and limitations
-
This study firstly identifies the circSCAPER/miR-140-3p/EZH2 axis OA process, gaining a new insight into the pathogenesis of OA.
-
However, the in vivo assay and a larger cohort of the disease analysis are essential to verify this conclusion.
Introduction
Osteoarthritis (OA) is one of the most common degenerative joint diseases, which is mainly characterized by joint inflammation, progressive destruction, and loss of articular cartilage, ultimately leading to severe joint pain, function loss, and physical disability.1,2 The loss of cartilage is usually the result of the imbalance of extracellular matrix (ECM) synthesis and catabolism as well as chondrocytes.3-5 Currently, existing evidence suggests that chondrocytes, the only cells in articular cartilage that produce and secrete ECM proteins to maintain cartilage integrity,6 play pivotal roles in the pathophysiology of OA through apoptosis and cytokine production.7,8 Thus, further studies on chondrocyte phenotype switching and ECM degradation might gain novel insights into the pathogenesis of OA.
Circular RNAs (circRNAs) are a kind of transcript with a covalently closed continuous loop that lacks the 5’-cap structure and the 3’-poly A tail, protecting them from the degradation of RNA exonuclease.9 CircRNAs are highly represented in the eukaryotic transcriptome, and show high stability, high evolutionary conservation between species, and tissue specificity.10 Growing evidence suggests that aberrant circRNA expression is related to the pathologies of diverse diseases through regulating biological processes such as proliferation, apoptosis, metabolism, and inflammation.11,12 Additionally, some circRNAs play critical roles in the development and progression of OA.13 For instance, circ_0136474,14 circRNA-UBE2G1,15 and circRNA.3318616 were found to accelerate the OA process through mediating chondrocyte phenotype switching. CircRNA S-phase cyclin A-associated protein in the ER (circSCAPER, ID: hsa_circ_0104595) is derived from the SCAPER gene and located at chr15: 77046148-77059429. SCAPER codes a cyclin A-interacting protein that regulates cell cycle progression.17,18 Mutations in the gene SCAPER have recently been shown to be implicated in causation of many types of diseases.19,20 A recent study discovered that hsa_circ_0104595 was highly expressed in synovial fluid of OA patients and had a high degree of specificity to serve as a biomarker for OA diagnosis; besides that, it was also associated with the severity of OA,21 suggesting that circSCAPER may play an important role in the pathogenesis of OA.
It has been revealed that interleukin (IL)-1β is abnormally overexpressed during OA progression, which could lead to the apoptosis of chondrocytes and degradation of articular ECM.22,23 Herein, this study used human normal chondrocyte C28/I2 that was exposed to IL-1β to induce cell injury to mimic the microenvironment of OA in vitro, then investigated the potential function and underlying pathogenic mechanisms of circSCAPER in OA.
Methods
Cartilage tissue samples
OA human cartilage tissues were isolated from 39 patients who received total knee arthroplasty surgery at Lhasa People's Hospital. The diagnosis of OA patients was based on guidelines from the American College of Rheumatology. The normal human cartilage tissues obtained from the femoral condyles and tibial plateaus of 39 donors during traumatic emergency amputation at the same hospital were used as controls, and no donors had a history of OA or rheumatoid arthritis. All cartilage tissues were immediately stored at -80°C. All enrolled individuals and their families were informed of the study and signed informed consent. The protocols of this work were permitted by the Ethics Committee of Lhasa People’s Hospital.
Cell culture and treatment
Human normal chondrocyte C28/I2 cell lines (Cell Bank of the Chinese Academy of Sciences, China) were cultured in a humidified atmosphere of 5% CO2 at 37°C with the RPMI-1640 medium (Gibco, USA) that contains 10% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin (Gibco). The C28/I2 chondrocytes were exposed to 10 ng/ml of IL-1β (MilliporeSigma, USA) for 24 hours to mimic the microenvironment of OA in vitro.
Quantitative real-time polymerase chain reaction
The nuclear and cytoplasmic RNAs of C28/I2 cells were isolated using the PARIS kit (Life Technologies, USA). Total RNA of cartilage tissue specimens and chondrocytes was extracted with TRIzol reagent (Life Technologies). Reverse transcription of the isolated total RNA was carried out using the Reverse Transcription Kit (Takara, Japan), and then SYBR green PCR Master Mix (Takara) was mixed with primers for quantitative real-time polymerase chain reaction (qRT-PCR) analysis. We employed the 2−ΔΔCT method to assess the relative fold changes of molecules. The same experiment was repeated three times, and the average fold change was taken. β-actin or U6 was used as the housekeeping gene for circSCAPER, enhancer of zeste homolog 2 (EZH2) or microRNA (miR)-140-3p, and cytoplasm and nucleus, respectively. The primers were listed as follows: circSCAPER: F 5’-GCTGAGTTTAAGCGAGAAGTGC-3’, R 5’- GCACTAGCAATGGCTTCTTCA-3’;
SCAPER: F 5’-AGGATGGGAGACCGTTCAGAG-3’, R 5’-AGAGGATGGTCACAAGAGTGT-3’; miR-140-3p: F 5’-GCCGAGTACCACAGGGTAGAA-3’, R 5’-CTCAACTGGTGTCGTGGA-3’;
β-actin: F 5’-GGCACTCTTCCAGCCTTCC-3’, R 5’-GAGCCGCCGATCCACAC-3’;
EZH2: F 5’-AGTGTTACCAGCATTTGGAGGG-3’, R 5’-CGGTGAGAGCAGCAGCAAAC-3’;
U6: F 5’-CTCGCTTCGGCAGCACA-3’, R 5’-AACGCTTCACGAATTTGCGT-3’.
RNase R treatment
Approximately 3 μg of RNA extracts were treated with or without 3 U/μg of RNase R (Qiagen, Japan) at 37°C for 15 minutes, then the resulting RNAs were purified employing RNeasy MinElute Cleanup Kit (Life Technologies). Finally, the abundance of circSCAPER and SCAPER messenger RNA (mRNA) was examined using qRT-PCR assay. The results represent the average of three independent replicates.
Cell transfection
The miR-140-3p mimic or inhibitor (miR-140-3p or in-miR-140-3p) and their negative control (miR-NC or in-miR-NC), circSCAPER-specific small interfering RNA (siRNA) (si-circSCAPER) and negative control (si-NC), pCD25-ciR-circSCAPER overexpression plasmid (circSCAPER) and plasmid empty control (NC), pcDNA 3.1-EZH2 overexpression plasmid (EZH2) and plasmid empty control (pcDNA) were obtained from Genepharma (China). The transfection was conducted in C28/I2 cells using Lipofectamine 2000 reagent (Invitrogen, Thermo Fisher Scientific, USA). After 48 hours of transfection, C28/I2 cells were treated with 10 ng/ml of IL-1β for 24 hours for subsequent analysis.
Western blot
Protein samples extracted from human cartilage tissues and cultured C28/I2 chondrocytes using the RIPA lysis buffer (Beyotime, China) were electrophoresed on 8% sodium dodecyl sulphate-polyacrylamide gel electrophoresis for separating. Then the samples were shifted onto the polyvinylidene fluoride (PVDF) membranes (Millipore, USA) and blocked in 5% non-fat milk for one hour. Thereafter, the membranes were incubated with specific primary antibody at 4°C overnight, followed by the secondary antibody for one hour at 37°C. The primary antibodies used in this study were as follows: anti-collagen II (1:1,000, ab188570), anti-matrix metallopeptidase 13 (MMP-13) (1:5,000, ab39012), anti-A Disintegrin and Metalloproteinase with Thrombospondin motifs (ADAMTS)-5 (1:500, ab41037), anti-B-cell lymphoma-2 (Bcl-2) (1:3,000, ab692), anti-Bcl-2-associated X (Bax) (1:3,000, ab32503), cleaved caspase 3 (1:3,000, ab2302), phosphoinositide 3-kinase (PI3K) (1:2,000, ab40776), phosphorylated(p)-PI3K (p-PI3K) (1:2,000, ab182651), and β-actin (1:1,000, ab6276), all obtained from Abcam (USA). Protein kinase B (AKT) (1:2,000, 9272) and p-AKT (1:2,000, 9271) were obtained from Cell Signaling Technology (USA). Experiments were performed three times. Finally, immunoreactive bands were measured employing the enhanced chemiluminescence solution (Beyotime).
Cell counting kit-8 assay
C28/I2 cells were transfected with assigned vectors, and then subjected to the treatment of IL-1β. After that, cells were seeded into each well of 96-well plates (1 × 104 cells/well), and incubated with 10 µl of cell counting kit-8 (CCK)-8 solution for two hours (5 mg/ml, MilliporeSigma). Then, optical value at 450 nm was detected at indicated times to assess cell proliferation. Each experiment was repeated three times independently.
Flow cytometry
Following indicated transfection and/or treatment, C28/I2 cells were resuspended in binding buffer (BD Biosciences, USA) to make the density at 1 × 106/ml, and then double-stained with 10 μl of annexin V-fluorescein isothiocyanate (FITC) and 10 μl of propidium iodide (PI) (BD Biosciences) for 15 minutes away from light. Finally, cell apoptosis was evaluated with the BD FACSCalibur flow cytometry (BD Biosciences) within one hour. Three replicate wells were set in each group, and the experiment was repeated three times.
Dual-luciferase reporter assay
The sequences of circSCAPER (position of 106-293) and EZH2 3' untranslated regions (3'UTR) (ID: ENSG00000106462, position of 379 -606), harboring the miR-140-3p seed regions or mutant sequences, were inserted into the pmirGLO luciferase vectors (GeneCreate, China) to generate circSCAPER wild-type (WT), circSCAPER mutant (MUT), EZH2 3’UTR WT, or EZH2 3’UTR MUT luciferase reporters. Then C28/I2 cells were transfected with these constructed luciferase report vectors and miR-140-3p mimic or miR-NC using Lipofectamine 2000 (Invitrogen). The dual luciferase reporter system (GeneCreate) was finally used to examine relative firefly luciferase activity. Each group was run in triplicate in 24-well plates.
RNA immunoprecipitation assay
C28/I2 cells were lysed in RNA immunoprecipitation (RIP) lysis buffer, and then the lysate was incubated with magnetic beads (Merck, Germany) coupled with human anti-Ago2 antibody (Abcam) and normal mouse immunoglobulin G (IgG) (Abcam). The magnetic beads were then incubated with the protease K buffer to remove the protein. The immunoprecipitated RNA was eluted and purified, and the levels of circSCAPER and miR-140-3p were examined using qRT-PCR assay. Triplicate individual experiments were performed in this study.
Statistical analysis
The results from three repeated experiments were presented as means and standard deviations (SDs) and were analyzed using GraphPad Prism 7 (GraphPad, USA). Differences in the level of gene expression were analyzed using independent-samples t-test. Pearson’s correlation coefficient analysis was used to assess the linear correlations. For in vitro experiments, the paired or unpaired t-test or one-way analysis of variance (ANOVA) was used to evaluate the differences between different groups. All p-values are two-tailed, and p < 0.05 indicates statistical significance.
Results
CircSCAPER expression is elevated in OA cartilage tissues
The expression profile of circSCAPER was first investigated using qRT-PCR. It was found that circSCAPER was highly expressed in OA cartilage tissues relative to the normal cartilage tissues (Figure 1a) (independent-samples t-test, p < 0.001). Besides that, the protein expression of cartilage matrix component (collagen II) and bone metabolic genes (MMP-13 and ADAMTS-5) was detected. In contrast with normal cartilage tissues, the levels of MMP-13 and ADAMTS-5 were elevated (independent-samples t-test, p < 0.001), while the level of collagen II was decreased (independent-samples t-test, p < 0.001) in OA cartilage tissues (Figure 1b), proving that circSCAPER was indeed highly expressed in OA patients. Then the stability of circSCAPER was determined, and circSCAPER was found to be prominently localized in the cytoplasm of C28/I2 chondrocytes (Figure 1c). Additionally, we found that circSCAPER was resistant to RNase R exonuclease in C28/I2 cells compared to the linear SCAPER mRNA (Figure 1d). Therefore, circSCAPER was abundant and stable in OA chondrocytes.
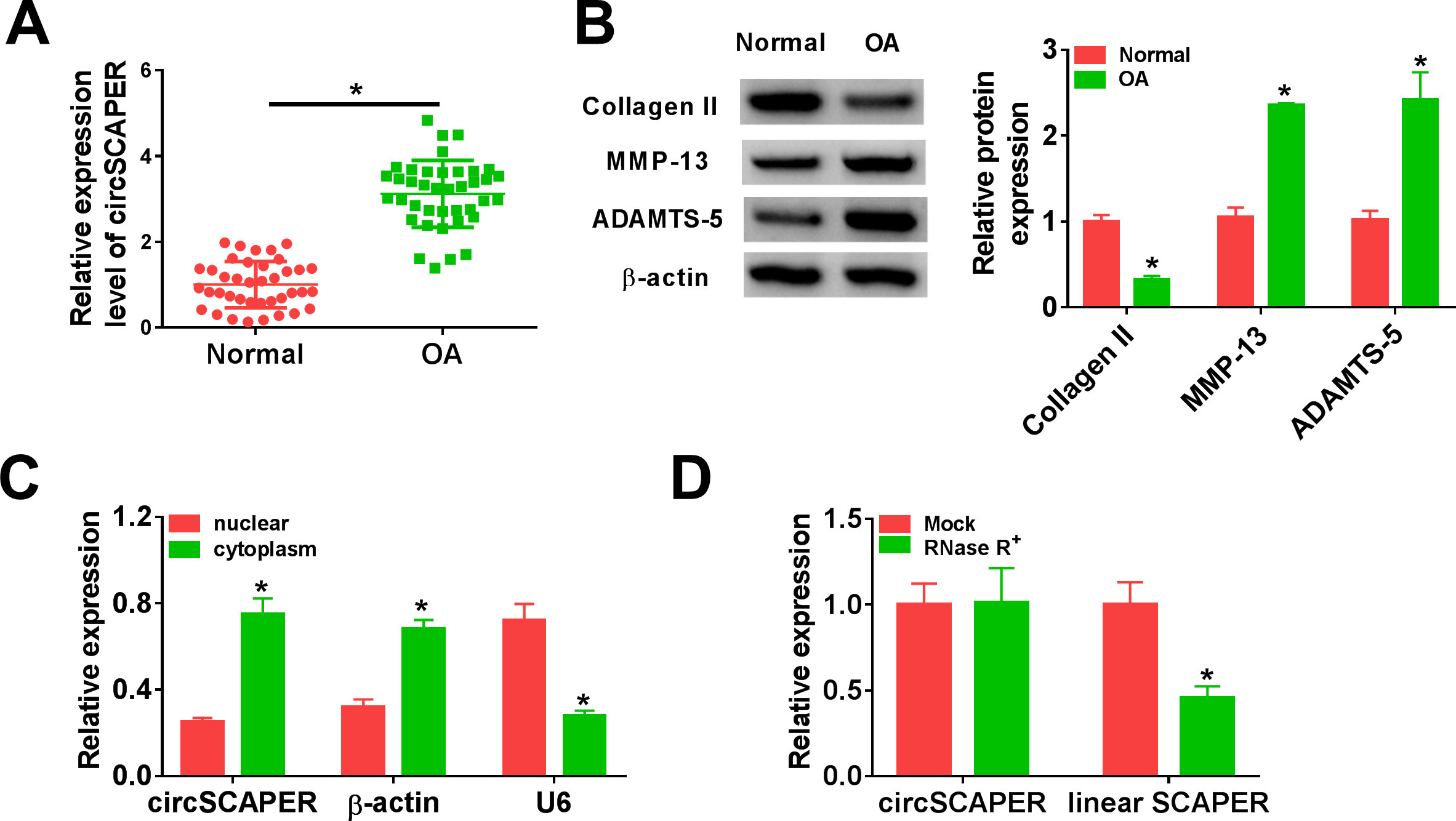
Fig. 1
CircSCAPER expression is elevated in osteoarthritis (OA) cartilage tissues. a) Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of circSCAPER expression level in OA cartilage tissues and normal cartilage tissues. b) Detection of matrix metallopeptidase 13 (MMP-13), a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS)-5, and collagen II protein levels in OA cartilage tissues using Western blot. c) qRT-PCR analysis of circSCAPER, β-actin, and U6 in purified C28/I2 nuclear and cytoplasmic fractions. d) qRT-PCR analysis of circSCAPER and SCAPER messenger RNA (mRNA) levels in C28/I2 cells treated with or without RNase R. Independent-samples t-test, *p < 0.05. All of the experiments were performed in triplicate.
CircSCAPER knockdown attenuates IL-1β-induced chondrocyte dysfunction
Next, the potential function of circSCAPER in OA was investigated. Cultured C28/I2 cells were exposed to IL-1β (10 ng/l) to induce an OA model in vitro. We found that IL-1β treatment elevated circSCAPER expression in C28/I2 cells (Figure 2a) (p < 0.001, independent-samples t-test). Then the siRNA targeting circSCAPER (si-circSCAPER) was constructed, and qRT-PCR analysis showed that transfection of si-circSCAPER reduced the level of circSCAPER in IL-1β-induced C28/I2 cells (Figure 2a) (p < 0.001, independent-samples t-test). Thereafter, we found that knockdown of circSCAPER attenuated IL-1β-induced elevation of MMP-13 and ADAMTS-5 levels (p < 0.001) and reduction of collagen II level (p = 0.014, both independent-samples t-test) (Figure 2b). CCK-8 and flow cytometry assays suggested that IL-1β suppressed the proliferation (p < 0.001) (Figure 2c) and induced apoptosis (p < 0.001) (Figure 2d) of C28/I2 cells, which were reversed by circSCAPER knockdown (p < 0.001, all independent-samples t-test). Moreover, apoptosis-related proteins were detected in IL-1β-induced C28/I2 cells, and Western blot analysis indicated that a decrease of circSCAPER upregulated Bcl-2 level (p < 0.001) but downregulated Bax (p < 0.001) and cleaved caspase 3 levels (p < 0.001, all independent-samples t-test) in cells (Figure 2e). Additionally, we analyzed the effects of circSCAPER on C28/I2 cells without IL-1β treatment. qRT-PCR analysis showed that transfection of si-circSCAPER remarkably reduced circSCAPER expression level in C28/I2 cells (Supplementary Figure aa). Further functional experiments showed that knockdown of circSCAPER alone did not affect cell ECM degradation, proliferation, and apoptosis (p > 0.050, independent-samples t-test) (Supplementary Figures ab to ae). Taken together, knockdown of circSCAPER reversed IL-1β-evoked inhibition of growth and degradation of ECM, thus impeding the process of OA.

Fig. 2
CircSCAPER knockdown attenuates interleukin (IL)-1β-induced chondrocyte dysfunction. a) to e) C28/I2 cells were transfected with circSCAPER-specific small interfering RNA (siRNA) (si-circSCAPER) and negative control (si-NC), followed by treatment with IL-1β (10 ng/l) for 24 hours. a) Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of circSCAPER expression in cells. b) Western blot analysis of matrix metallopeptidase 13 (MMP-13), a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS-5), and collagen II protein levels in cells. c) Cell counting kit-8 (CCK-8) assay for cell proliferation. d) Flow cytometry for cell apoptosis. e) Western blot analysis of B-cell lymphoma-2 (Bcl-2), Bcl-2-associated X (Bax), and cleaved caspase 3 protein levels in cells. One-way analysis of variance (ANOVA), *p < 0.05. All of the experiments were performed in triplicate.
MiR-140-3p is a target of circSCAPER
Given that circRNAs could act as miRNA sponges in the cytoplasm, and circSCAPER was prominently localized in the cytoplasm of C28/I2 chondrocytes, the potential targets of circSCAPER were firstly predicted by searching the CircInteractome online database,24,25 and we found that circSCAPER possesses conserved target site of miR-140-3p with a high score (Figure 3a). Then dual-luciferase reporter assay was applied in C28/I2 cells, and we found that overexpression of miR-140-3p significantly reduced the luciferase activity of the WT group (p < 0.001) but not of the MUT group (p = 0.788, both independent-samples t-test) (Figure 3B). Furthermore, RIP assay suggested that circSCAPER and miR-140-3p were efficiently pulled down by anti-Ago2 antibodies relative to IgG antibodies (p < 0.001, independent-samples t-test) (Figure 3c). All these data confirmed the binding of circSCAPER and miR-140-3p. Next, the expression of miR-140-3p in OA was investigated. miR-140-3p expression was lower in OA cartilage tissues than that in normal cartilage tissues (p < 0.001) (Figure 3d), which was negatively correlated with circSCAPER expression (p < 0.001, both independent-samples t-test) (Figure 3e). Also, miR-140-3p expression was decreased in IL-1β-induced C28/I2 cells (p = 0.002, independent-samples t-test) (Figure 3f). Besides that, after confirming the transfection of circSCAPER overexpression vector using qRT-PCR (p = 0.002) (Figure 3g), it was proved that circSCAPER overexpression suppressed miR-140-3p expression in C28/I2 cells (p = 0.004), while its downregulation elevated miR-140-3p expression (p < 0.001, all independent-samples t-test) (Figure 3h). In all, we confirmed that circSCAPER directly bound to miR-140-3p and negatively regulated its expression.

Fig. 3
MicroRNA (miR)-140-3p is a target of circSCAPER. a) The potential binding sites between circSCAPER and miR-140-3p. b) The relative luciferase activity analysis in C28/I2 cells after transfection with circSCAPER wild-type (WT) or circSCAPER mutant (MUT) and miR-140-3p mimics or miR-negative control (NC) using dual-luciferase reporter assay. c) Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of circSCAPER and miR-140-3p levels in C28/I2 cells after RNA immunoprecipitation (RIP) assay. d) qRT-PCR analysis of miR-140-3p expression level in osteoarthritis (OA) cartilage tissues and normal cartilage tissues. e) Correlation analysis between miR-140-3p and circSCAPER expression in OA cartilage tissues. f) Detection of miR-140-3p level in C28/I2 cells treated with interleukin (IL)-1β (10 ng/l). g) Detection of circSCAPER level in C28/I2 cells transfected with circSCAPER or NC. h) qRT-PCR analysis of miR-140-3p expression level in C28/I2 cells transfected with circSCAPER, NC, circSCAPER-specific small interfering RNA (siRNA) (si-circSCAPER), or si-NC. Independent-samples t-test, *p < 0.05. All of the experiments were performed in triplicate. IgG, immunoglobulin G.
CircSCAPER knockdown attenuates IL-1β-induced chondrocyte dysfunction via miR-140-3p
We then elucidated whether miR-140-3p mediated the effects of circSCAPER on the OA process. C28/I2 cells were co-transfected with si-circSCAPER and/or in-miR-140-3p, followed by IL-1β (10 ng/l) treatment for 24 hours, then we found that inhibition of miR-140-3p reduced si-circSCAPER-induced elevation of miR-140-3p level in IL-1β-induced C28/I2 cells (p < 0.001, one-way ANOVA) (Figure 4a). Afterwards, it was proved that miR-140-3p inhibition abolished circSCAPER knockdown-evoked repression of ECM degradation (p < 0.001) (Figure 4b), promotion of cell proliferation (p < 0.001) (Figure 4c), and arrest of cell apoptosis (p < 0.001, all one-way ANOVA) (Figures 4d and 4e) in IL-1β-induced C28/I2 cells. Therefore, we demonstrated that circSCAPER/miR-140-3p axis might be responsible for IL-1β-induced OA.
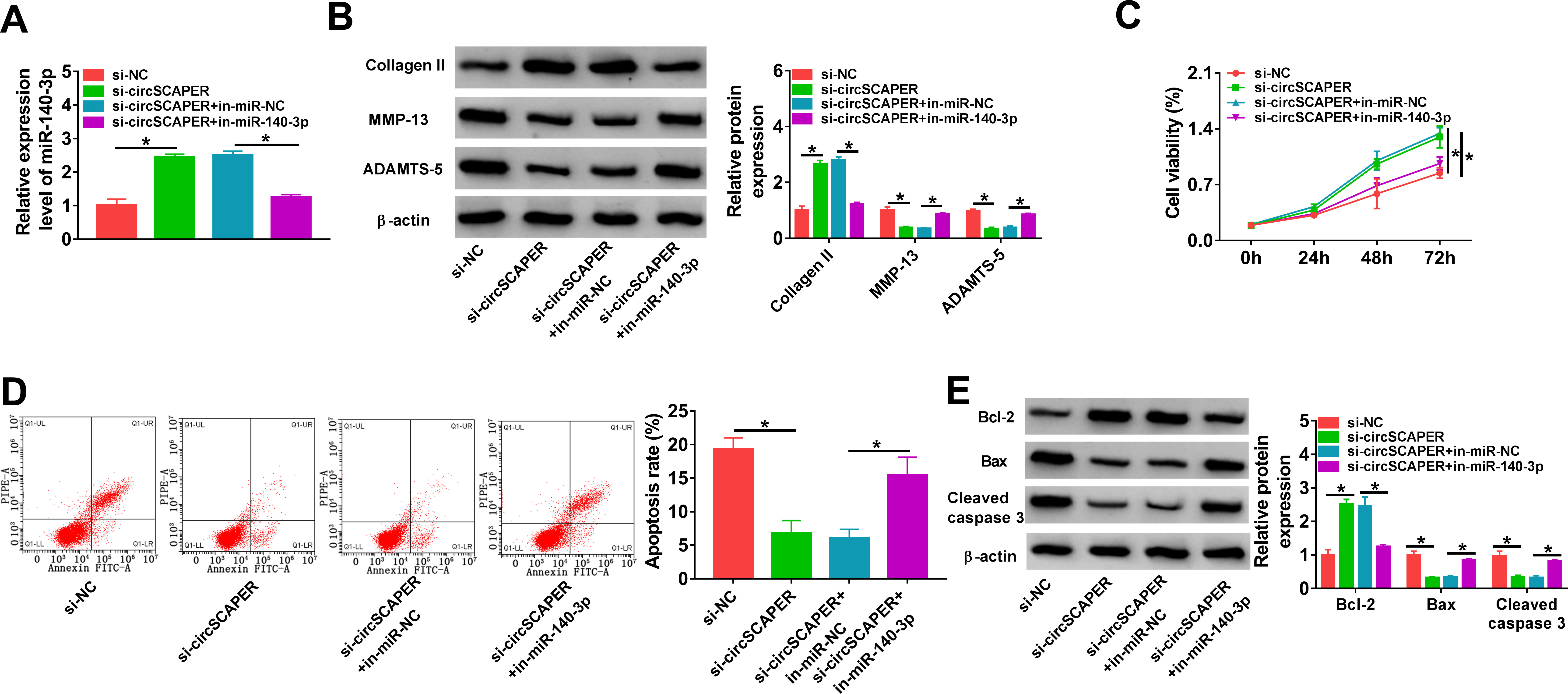
Fig. 4
CircSCAPER knockdown attenuates interleukin (IL)-1β-induced chondrocyte dysfunction via microRNA (miR)-140-3p. a) to e) C28/I2 cells were transfected with circSCAPER-specific small interfering RNA (siRNA) (si-circSCAPER), si-negative control (NC), si-circSCAPER+in-miR-NC, or si-circSCAPER+in-miR-140-3p, followed by treatment with IL-1β (10 ng/l) for 24 hours. a) Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of miR-140-3p level in cells. b) Western blot analysis of matrix metallopeptidase 13 (MMP-13), a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS)-5, and collagen II protein levels in cells. c) Cell counting kit-8 (CCK-8) assay for cell proliferation. d) Flow cytometry for cell apoptosis. e) Western blot analysis of B-cell lymphoma-2 (Bcl-2), Bcl-2-associated X (Bax), and cleaved caspase 3 protein levels in cells. One-way analysis of variance (ANOVA), *p < 0.05. All of the experiments were performed in triplicate. NC, negative control.
CircSCAPER functions as a sponge for miR-140-3p to regulate the expression of its target EZH2
The molecular mechanism underlying miR-140-3p was further elucidated in our study. According to the prediction of DIANA tool-microT_CDS software, a potential binding site existed between miR-140-3p and EZH2 (Figure 5a). Further dual-luciferase reporter assay suggested that miR-140-3p mimics remarkably decreased the luciferase activity of the wild-type EZH2 reporter compared with the control group (p = 0.001), while no change was observed in mutant EZH2 reporter after overexpression of miR-140-3p (p = 0.909, both independent-samples t-test) (Figure 5b), confirming the direct interaction between miR-140-3p and EZH2. After that, the expression profile of EZH2 was investigated. By contrast with normal cartilage tissues, EZH2 expression was higher in OA cartilage tissues at mRNA (p < 0.001) and protein (p < 0.001) levels (Figures 5c and 5d), which was negatively correlated with miR-140-3p expression (p < 0.001) (Figure 5e) and positively correlated with circSCAPER expression (p < 0.001, all independent-samples t-test) (Figure 5f). In addition, we discovered that IL-1β treatment elevated EZH2 expression (p < 0.001, independent-samples t-test) (Figure 5g). After confirming the transfection efficiency of miR-140-3p mimic (p < 0.001) or inhibitor (p = 0.009) (Figure 5h), it was confirmed that miR-140-3p overexpression suppressed EZH2 expression (p < 0.001) (Figure 5i); importantly, knockdown of circSCAPER reduced EZH2 expression (p = 0.001), which was rescued by miR-140-3p inhibition (p < 0.001, all independent-samples t-test) (Figure 5j). Therefore, we verified that miR-140-3p directly targeted EZH2 and negatively regulated its expression; besides that, circSCAPER could indirectly regulate EZH2 expression via sponging miR-140-3p.

Fig. 5
CircSCAPER functions as a sponge for microRNA (miR)-140-3p to regulate the expression of its target enhancer of zeste homolog 2 (EZH2). a) The potential binding sites between miR-140-3p and EZH2. b) The analysis of relative luciferase activity in C28/I2 cells after transfection with EZH2 3' untranslated region (3'UTR) wild-type (WT) or EZH2 3’UTR mutant (MUT) and miR-140-3p mimics or miR-negative control (NC) using dual-luciferase reporter assay. c) and d) Quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot analysis of EZH2 level in osteoarthritis (OA) cartilage tissues. e) and f) Correlation analysis between miR-140-3p and EZH2 messenger RNA (mRNA) or circSCAPER expression in OA cartilage tissues. g) Measurement of EZH2 protein level in C28/I2 cells treated with interleukin (IL)-1β (10 ng/l). h) qRT-PCR analysis of miR-140-3p expression level in C28/I2 cells transfected with in-miR-NC, in-miR-140-3p, miR-NC, or miR-140-3p. i) Western blot analysis of EZH2 level in C28/I2 cells transfected with miR-NC or miR-140-3p. j) Western blot analysis of EZH2 level in C28/I2 cells transfected with circSCAPER-specific small interfering RNA (siRNA) (si-circSCAPER), si-NC, si-circSCAPER+ in-miR-NC, or si-circSCAPER+ in-miR-140-3p. Independent-samples t-test, *p < 0.05. All of the experiments were performed in triplicate. IgG, immunoglobulin G.
miR-140-3p protects chondrocytes against IL-1β-induced dysfunction through EZH2
To explore the function of the miR-140-3p/EZH2 axis in regulating the OA process, miR-140-3p and/or EZH2 were transfected into C28/I2 cells, then cells were treated with IL-1β (10 ng/l) for 24 hours. Western blot analysis showed that miR-140-3p reduced the level of EZH2 (p < 0.001), which was rescued by the transfection of EZH2 overexpression vector in IL-1β-induced C28/I2 cells (p < 0.001, both one-way ANOVA) (Figure 6a). Thereafter, we found that miR-140-3p re-expression in IL-1β-induced C28/I2 cells elevated the expression of collagen II and reduced the levels of MMP-13 and ADAMTS-5 (p < 0.001), which was reverted by EZH2 upregulation (p < 0.001, both one-way ANOVA) (Figure 6b). Besides that, it was proved that restoration of miR-140-3p promoted proliferation (p < 0.001) (Figure 6c) and suppressed apoptosis (p = 0.001) (Figures 6d and 6e) in IL-1β-induced C28/I2 cells, while this condition was abolished by EZH2 overexpression (p < 0.001, all one-way ANOVA) (Figures 6c and 6e). Collectively, miR-140-3p could impair IL-1β-triggered ECM degradation and apoptosis in IL-1β-induced chondrocytes via EZH2, suggesting that the miR-140-3p/EZH2 axis was engaged in the OA process. Besides,

Fig. 6
MicroRNA (miR)-140-3p protects chondrocytes against interleukin (IL)-1β-induced dysfunction through enhancer of zeste homolog 2 (EZH2). a) to e) C28/I2 cells were transfected with miR-NC, miR-140-3p, miR-140-3p+pcDNA, or miR-140-3p+EZH2, followed by treatment with IL-1β (10 ng/l) for 24 hours. a) Western blot analysis of EZH2 protein level in cells. b) Western blot analysis of matrix metallopeptidase 13 (MMP-13), a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS)-5, and collagen II protein levels in cells. c) Cell counting kit-8 (CCK-8) assay for cell proliferation. d) Flow cytometry for cell apoptosis. e) Detection of B-cell lymphoma-2 (Bcl-2), Bcl-2-associated X (Bax), and cleaved caspase 3 protein levels in cells using Western blot. One-way analysis of variance (ANOVA), *p < 0.05. All of the experiments were performed in triplicate.
CircSCAPER can activate the PI3K/AKT pathway through miR-140-3p/EZH2 axis
The PI3K/AKT pathway is a pivotal regulator of normal cellular processes involved in cell proliferation, motility, apoptosis, survival, and metabolism. Therefore, we wanted to study whether the circSCAPER/miR-140-3p/EZH2 axis could exert its effects via the PI3K/AKT pathway. As shown in Figures 7a and 7b, we found that knockdown of circSCAPER reduced the levels of p-AKT and p-PI3K in IL-1β-induced C28/I2 cells (p < 0.001), which was rescued by miR-140-3p inhibition (p < 0.001, both one-way ANOVA). Meanwhile, miR-140-3p overexpression was also found to decrease p-AKT and p-PI3K levels (p < 0.001), while EZH2 overexpression could abolish this condition (p < 0.001, both one-way ANOVA). These data suggest that circSCAPER could activate the PI3K/AKT pathway through the miR-140-3p/EZH2 axis. Based on the aforementioned, circSCAPER promoted IL-1β-stimulated ECM degradation, proliferation arrest, and apoptosis enhancement in chondrocytes via regulating the miR-140-3p/EZH2 axis (Figure 8).

Fig. 7
CircSCAPER can activate the phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT) pathway through the microRNA (miR)-140-3p/enhancer of zeste homolog 2 (EZH2) axis. a) Western blot analysis of p-AKT and p-PI3K levels in C28/I2 cells transfected with circSCAPER-specific small interfering RNA (siRNA) (si-circSCAPER), si-negative control (NC), si-circSCAPER+in-miR-NC, or si-circSCAPER+in-miR-140-3p in the presence of interleukin (IL)-1β. b) Western blot analysis of p-AKT and p-PI3K levels in C28/I2 cells transfected with miR-NC, miR-140-3p, miR-140-3p+pcDNA, or miR-140-3p+EZH2 in the presence of IL-1β. One-way analysis of variance, *p < 0.05. All of the experiments were performed in triplicate.
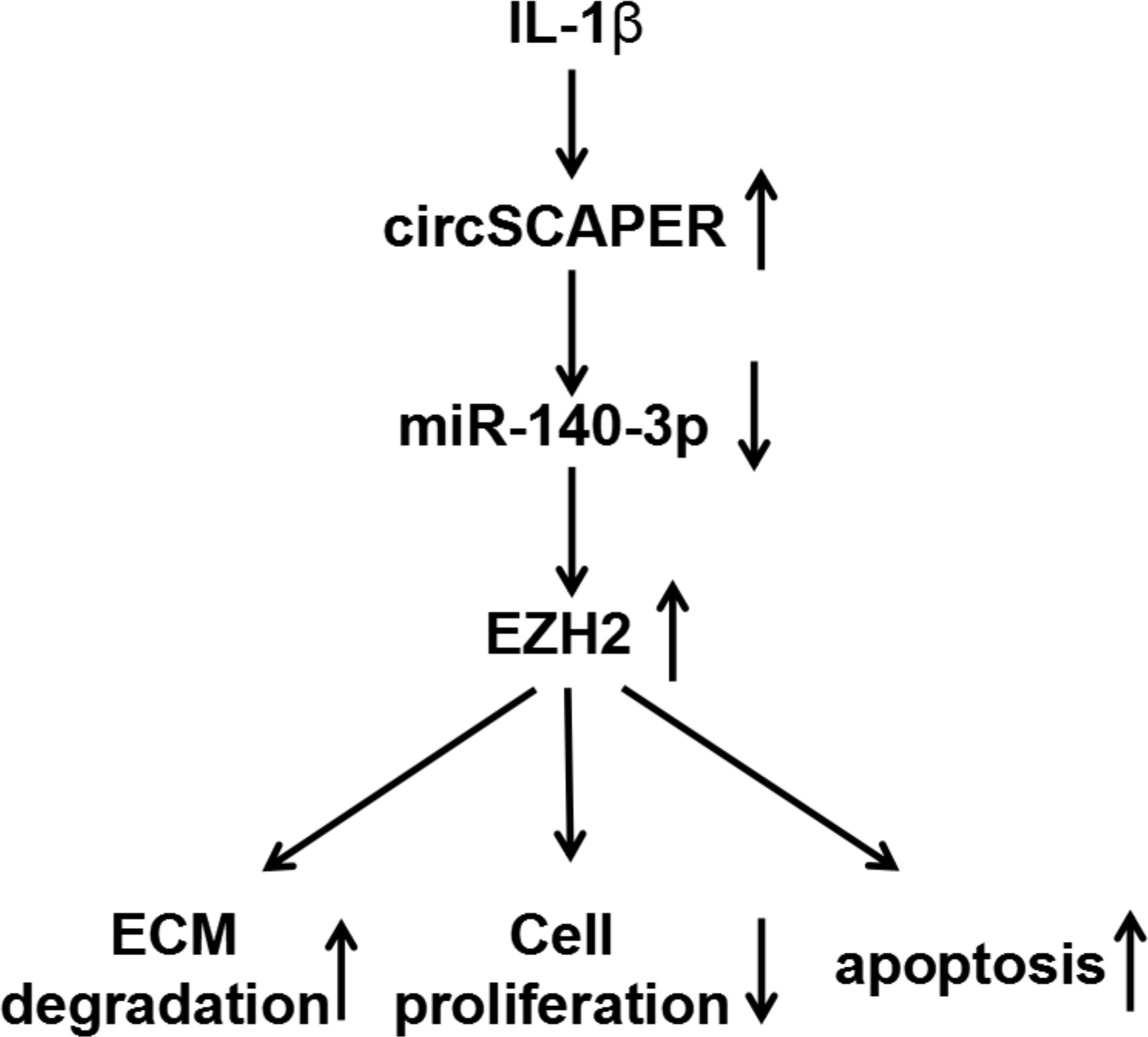
Fig. 8
Graphical abstract of how circSCAPER promotes interleukin (IL)-1β-induced chondrocyte dysfunction. CircSCAPER promotes IL-1β-stimulated extracellular matrix (ECM) degradation, proliferation arrest, and apoptosis enhancement in chondrocytes via regulating the microRNA (miR)-140-3p/enhancer of zeste homolog 2 (EZH2) axis.
Discussion
CircRNAs are widely and abundantly expressed in mammalian cells and stably presented in intracellular and humoral circulation compared with other non-coding RNAs, such as long non-coding RNAs (lncRNAs) and miRNAs.26 These properties render circRNAs to be potential applicable biomarkers in routine clinical practice for human diseases screening. Recently, the role of circRNA in OA progression has attracted great research interest. Numerous circRNAs were found to be differentially expressed in pathological status of OA.27 Functionally, circRNA participates in regulating the dysfunction of chondrocytes, in terms of proliferation and apoptosis, and the imbalance of ECM homeostasis, thus affecting OA development.28-30 In the current study, circSCAPER was found to be elevated in OA cartilage tissues. Then cultured human normal chondrocytes were treated with IL-1β to mimic the microenvironment of OA in vitro. As expected, circSCAPER expression was also elevated in IL-1β-induced chondrocytes. Furthermore, knockdown of circSCAPER in IL-1β-induced chondrocytes reduced cell apoptosis and induced cell proliferation. Collagen II is the major collagen component in the cartilage matrix, while MMP-13 and ADAMTS-5 are identified as major cartilage-degrading enzymes that break down components of the ECM.31 In vitro assay also showed that MMP-13 and ADAMTS-5 levels were decreased, while collagen II level was increased in IL-1β-induced chondrocytes after circSCAPER reduction. Therefore, we demonstrated that circSCAPER silencing promoted cell proliferation and ECM formation, suggesting that circSCAPER is associated with the progression of OA.
The competing endogenous RNAs (ceRNAs) hypothesis proposes that circRNAs interact with miRNAs by acting as ceRNAs to regulate cellular physiological function.32,33 miRNAs have been shown to be closely related to the progression and severity of OA.34,35 Therefore, the regulatory mechanism underlying circSCAPER in OA was further explored, and we confirmed that circSCAPER directly binds to miR-140-3p. miR-140-3p isomiRs are abundantly expressed in cartilage, which is more effective than the original miR-140-3p annotated in miRBase36-38 in human and murine chondrocytes,39 suggesting the potential function of miR-140-3p in OA. Besides that, it was also demonstrated that miR-140-3p was downregulated in OA, and negatively correlated with OA severity; furthermore, miR-140-3p re-expression alleviated OA progression.40,41 In this study, we also validated a decrease of miR-140-3p expression in OA cartilage tissues and IL-1β-induced chondrocytes; moreover, miR-140-3p upregulation induced impairment of cell apoptosis and ECM degradation as well as promotion of cell proliferation in IL-1β-induced chondrocytes. Importantly, inhibition of miR-140-3p abrogated the protective effects of circSCAPER knockdown on chondrocytes. Additionally, this study investigated the downstream gene of miR-140-3p. We verified that miR-140-3p directly targeted EZH2; furthermore, circSCAPER acted as a sponge for miR-140-3p to positively regulate EZH2 expression. EZH2, the enzymatic component of polycomb repressive complex 2 (PRC2), is an evolutionary conserved gene. It has been found to be aberrantly overexpressed in a wide range of malignancies, and functions as an oncogene to promote cancer progression.42,43 In addition, EZH2 expression was significantly elevated in the chondrocytes of OA patients compared to normal humans,44 and inhibition of EZH2 reduced cartilage loss and functional impairment related to OA by decreasing the expression of genes involved in inflammation, pain (nitric oxide (NO), prostaglandin E2 (PGE2), nerve growth factor (NGF), interleukin-6 (IL-6)), and catabolism (MMPs).45 Chen et al46 found that EZH2 could activate Wnt/β-catenin signalling to upregulate the expression of catabolic genes collagen X (COLX) and ADAMTS-5, contributing to the degradation of cartilage-specific ECM and chondrocyte hypertrophy.46 Li et al suggested that EZH2 promoted ECM degradation and apoptosis in OA chondrocytes.47 In this study, EZH2 was also elevated in OA cartilage tissues and cells, and overexpression of EZH2 abolished the effects of miR-140-3p on IL-1β-stimulated chondrocytes.
The PI3K/AKT pathway is implicated in modulating cell proliferation, migration, adhesion, death, and tumorigenesis in a variety of cell lineages.48 However, PI3K/Akt signalling shows pleiotropic functions in chondrocytes in OA. On the one hand, some studies showed that activation of this pathway by insulin-like growth factor 1 (IGF-1) could boost chondrocyte survival as well as cartilage matrix synthesis.49,50 On the other hand, when this pathway is activated by inflammatory cytokines, such as IL-1β, it might induce the generation of MMPs, leading to the loss of cartilage matrix.51,52 In the current study, we found that circSCAPER could enhance the phosphorylation of PI3K and AKT via the miR-140-3p/EZH2 axis, thus resulting in activation of the PI3K/AKT pathway. However, the detailed role of the PI3K/AKT pathway in this study still needs further investigation.
In conclusion, this work uncovered that circSCAPER promoted IL-1β-stimulated ECM degradation, proliferation arrest, and apoptosis enhancement in chondrocytes via regulating the miR-140-3p/EZH2 axis, suggesting a novel insight into OA pathogenesis and the development of an effective therapeutic approach for OA.
References
1. Glyn-Jones S , Palmer AJR , Agricola R , et al. Osteoarthritis . Lancet . 2015 ; 386 ( 9991 ): 376 – 387 . Crossref PubMed Google Scholar
2. He Z , Nie P , Lu J , et al. Less mechanical loading attenuates osteoarthritis by reducing cartilage degeneration, subchondral bone remodelling, secondary inflammation, and activation of NLRP3 inflammasome . Bone Joint Res . 2020 ; 9 ( 10 ): 731 – 741 . Crossref PubMed Google Scholar
3. Wang Q , Rozelle AL , Lepus CM , et al. Identification of a central role for complement in osteoarthritis . Nat Med . 2011 ; 17 ( 12 ): 1674 – 1679 . Crossref PubMed Google Scholar
4. Chen D , Shen J , Zhao W , et al. Osteoarthritis: toward a comprehensive understanding of pathological mechanism . Bone Res . 2017 ; 5 : 16044 . Crossref PubMed Google Scholar
5. Akhbari P , Karamchandani U , Jaggard MKJ , et al. Can joint fluid metabolic profiling (or “metabonomics”) reveal biomarkers for osteoarthritis and inflammatory joint disease? Bone Joint Res . 2020 ; 9 ( 3 ): 108 – 119 . Google Scholar
6. Woods A , Wang G , Beier F . Regulation of chondrocyte differentiation by the actin cytoskeleton and adhesive interactions . J Cell Physiol . 2007 ; 213 ( 1 ): 1 – 8 . Crossref PubMed Google Scholar
7. Aigner T , Söder S , Gebhard PM , McAlinden A , Haag J . Mechanisms of disease: role of chondrocytes in the pathogenesis of osteoarthritis--structure, chaos and senescence . Nat Clin Pract Rheumatol . 2007 ; 3 ( 7 ): 391 – 399 . Crossref PubMed Google Scholar
8. Woodell-May JE , Sommerfeld SD . Role of inflammation and the immune system in the progression of osteoarthritis . J Orthop Res . 2020 ; 38 ( 2 ): 253 – 257 . Crossref PubMed Google Scholar
9. Barrett SP , Wang PL , Salzman J . Role of inflammation and the immune system in the progression of osteoarthritis . Elife . 2015 ; 4 : e07540 . Crossref PubMed Google Scholar
10. Jeck WR , Sorrentino JA , Wang K , et al. Circular RNAs are abundant, conserved, and associated with Alu repeats . RNA . 2013 ; 19 ( 2 ): 141 – 157 . Crossref PubMed Google Scholar
11. Yu C-Y , Kuo H-C . The emerging roles and functions of circular RNAs and their generation . J Biomed Sci . 2019 ; 26 ( 1 ): 29 . Crossref PubMed Google Scholar
12. Marques-Rocha JL , Samblas M , Milagro FI , Bressan J , Martínez JA , Marti A . Noncoding RNAs, cytokines, and inflammation-related diseases . FASEB J . 2015 ; 29 ( 9 ): 3595 – 3611 . Crossref PubMed Google Scholar
13. Li H , Yang HH , Sun ZG , Tang HB , Min JK . Whole-transcriptome sequencing of knee joint cartilage from osteoarthritis patients . Bone Joint Res . 2019 ; 8 ( 7 ): 290 – 303 . Crossref PubMed Google Scholar
14. Li Z , Yuan B , Pei Z , et al. Circ_0136474 and MMP-13 suppressed cell proliferation by competitive binding to miR-127-5p in osteoarthritis . J Cell Mol Med . 2019 ; 23 ( 10 ): 6554 – 6564 . Crossref PubMed Google Scholar
15. Chen G , Liu T , Yu B , Wang B , Peng Q . CircRNA-UBE2G1 regulates LPS-induced osteoarthritis through miR-373/HIF-1a axis . Cell Cycle . 2020 ; 19 ( 13 ): 1696 – 1705 . Crossref PubMed Google Scholar
16. Zhou Z-B , Huang G-X , Fu Q , et al. circRNA.33186 contributes to the pathogenesis of osteoarthritis by sponging miR-127-5p . Mol Ther . 2019 ; 27 ( 3 ): 531 – 541 . Crossref PubMed Google Scholar
17. Tsang WY , Wang L , Chen Z , Sánchez I , Dynlacht BD . SCAPER, a novel cyclin A-interacting protein that regulates cell cycle progression . J Cell Biol . 2007 ; 178 ( 4 ): 621 – 633 . Crossref PubMed Google Scholar
18. Richards S , Aziz N , Bale S , et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology . Genet Med . 2015 ; 17 ( 5 ): 405 – 423 . Crossref PubMed Google Scholar
19. Jauregui R , Thomas AL , Liechty B , et al. SCAPER-associated nonsyndromic autosomal recessive retinitis pigmentosa . Am J Med Genet A . 2019 ; 179 ( 2 ): 312 – 316 . Crossref PubMed Google Scholar
20. Tatour Y , Sanchez-Navarro I , Chervinsky E , et al. Mutations in SCAPER cause autosomal recessive retinitis pigmentosa with intellectual disability . J Med Genet . 2017 ; 54 ( 10 ): 698 – 704 . Crossref PubMed Google Scholar
21. Yu F , Xie C , Sun J , Feng H , Huang X . Circular RNA expression profiles in synovial fluid: a promising new class of diagnostic biomarkers for osteoarthritis . Int J Clin Exp Pathol . 2018 ; 11 ( 3 ): 1338 – 1346 . PubMed Google Scholar
22. Kapoor M , Martel-Pelletier J , Lajeunesse D , Pelletier J-P , Fahmi H . Role of proinflammatory cytokines in the pathophysiology of osteoarthritis . Nat Rev Rheumatol . 2011 ; 7 ( 1 ): 33 – 42 . Crossref PubMed Google Scholar
23. Pelletier JP , Faure MP , DiBattista JA , Wilhelm S , Visco D , Martel-Pelletier J . Coordinate synthesis of stromelysin, interleukin-1, and oncogene proteins in experimental osteoarthritis. An immunohistochemical study . Am J Pathol . 1993 ; 142 ( 1 ): 95 – 105 . PubMed Google Scholar
24. No authors listed . Circular RNA Interactome . National Institute of Aging, National Institutes of Health . 2020 . https://circinteractome.irp.nia.nih.gov ( date last accessed 19 January 2022 ). Google Scholar
25. Dudekula DB , Panda AC , Grammatikakis I , De S , Abdelmohsen K , Gorospe M . CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs . RNA Biol . 2016 ; 13 ( 1 ): 34 – 42 . Crossref PubMed Google Scholar
26. Geng Y , Jiang J , Wu C . Function and clinical significance of circRNAs in solid tumors . J Hematol Oncol . 2018 ; 11 ( 1 ): 98 . Crossref PubMed Google Scholar
27. Wang Y , Wu C , Zhang Y , et al. Screening for differentially expressed circRNA between Kashin–Beck disease and osteoarthritis patients based on circRNA chips . Clinica Chimica Acta . 2020 ; 501 ( 11 ): 92 – 101 . Google Scholar
28. Zhou Z-B , Du D , Huang G-X , Chen A , Zhu L . Circular RNA Atp9b, a competing endogenous RNA, regulates the progression of osteoarthritis by targeting miR-138-5p . Gene . 2018 ; 646 ( 9783 ): 203 – 209 . Crossref PubMed Google Scholar
29. Wu Y , Zhang Y , Zhang Y , Wang J-J . CircRNA hsa_circ_0005105 upregulates NAMPT expression and promotes chondrocyte extracellular matrix degradation by sponging miR-26a . Cell Biol Int . 2017 ; 41 ( 12 ): 1283 – 1289 . Crossref PubMed Google Scholar
30. Li B-F , Zhang Y , Xiao J , et al. Hsa_circ_0045714 regulates chondrocyte proliferation, apoptosis and extracellular matrix synthesis by promoting the expression of miR-193b target gene IGF1R . Hum Cell . 2017 ; 30 ( 4 ): 311 – 318 . Crossref PubMed Google Scholar
31. Zhang Y , Wang F , Chen G , He R , Yang L . LncRNA MALAT1 promotes osteoarthritis by modulating miR-150-5p/AKT3 axis . Cell Biosci . 2019 ; 9 ( 1 ): 54 . Crossref PubMed Google Scholar
32. Hansen TB , Jensen TI , Clausen BH , et al. Natural RNA circles function as efficient microRNA sponges . Nature . 2013 ; 495 ( 7441 ): 384 – 388 . Crossref PubMed Google Scholar
33. Thomson DW , Dinger ME . Endogenous microRNA sponges: evidence and controversy . Nat Rev Genet . 2016 ; 17 ( 5 ): 272 – 283 . Crossref PubMed Google Scholar
34. Zhang A , Ma S , Yuan L , et al. Knockout of miR-21-5p alleviates cartilage matrix degradation by targeting Gdf5 in temporomandibular joint osteoarthritis . Bone Joint Res . 2020 ; 9 ( 10 ): 689 – 700 . Crossref PubMed Google Scholar
35. Brzeszczyńska J , Brzeszczyński F , Hamilton DF , McGregor R , Simpson AHRW . Role of microRNA in muscle regeneration and diseases related to muscle dysfunction in atrophy, cachexia, osteoporosis, and osteoarthritis . Bone Joint Res . 2020 ; 9 ( 11 ): 798 – 807 . Crossref PubMed Google Scholar
36. No authors listed . miRBase: the microRNA database . The University of Manchester . 2022 . https://www.mirbase.org ( date last accessed 19 January 2022 ). Google Scholar
37. Kozomara A , Griffiths-Jones S . miRBase: annotating high confidence microRNAs using deep sequencing data . Nucleic Acids Res . 2014 ; 42 ( Database issue ): D68 - 73 . Crossref PubMed Google Scholar
38. Kozomara A , Birgaoanu M , Griffiths-Jones S . miRBase: from microRNA sequences to function . Nucleic Acids Res . 2019 ; 47 ( D1 ): D155 – D162 . Crossref PubMed Google Scholar
39. Woods S , Charlton S , Cheung K , et al. microRNA-seq of cartilage reveals an overabundance of miR-140-3p which contains functional isomiRs . RNA . 2020 ; 26 ( 11 ): 1575 – 1588 . Crossref PubMed Google Scholar
40. Yin C-M , Suen W-C-W , Lin S , Wu X-M , Li G , Pan X-H . Dysregulation of both miR-140-3p and miR-140-5p in synovial fluid correlate with osteoarthritis severity . Bone Joint Res . 2017 ; 6 ( 11 ): 612 – 618 . Crossref PubMed Google Scholar
41. Ren T , Wei P , Song Q , Ye Z , Wang Y , Huang L . MiR-140-3p Ameliorates the Progression of Osteoarthritis via Targeting CXCR4 . Biol Pharm Bull . 2020 ; 43 ( 5 ): 810 – 816 . Crossref PubMed Google Scholar
42. Gan L , Yang Y , Li Q , Feng Y , Liu T , Guo W . Epigenetic regulation of cancer progression by EZH2: from biological insights to therapeutic potential . Biomarker Research . 2018 ; 6 ( 1 ): 10 . Crossref PubMed Google Scholar
43. Wang T , Wang X , Du Q , et al. The circRNA circP4HB promotes NSCLC aggressiveness and metastasis by sponging miR-133a-5p . Biochem Biophys Res Commun . 2019 ; 513 ( 4 ): 904 – 911 . Crossref PubMed Google Scholar
44. He J , Cao W , Azeem I , Shao Z . Epigenetics of osteoarthritis: histones and TGF-β1 . Clinica Chimica Acta . 2020 ; 510 ( 10 ): 593 – 598 . Crossref PubMed Google Scholar
45. Allas L , Brochard S , Rochoux Q , et al. EZH2 inhibition reduces cartilage loss and functional impairment related to osteoarthritis . Sci Rep . 2020 ; 10 ( 1 ): 19577 . Crossref PubMed Google Scholar
46. Chen L , Wu Y , Wu Y , Wang Y , Sun L , Li F . The inhibition of EZH2 ameliorates osteoarthritis development through the Wnt/β-catenin pathway . Sci Rep . 2016 ; 6 ( 1 ): 29176 . Crossref PubMed Google Scholar
47. Li Y , Yuan F , Song Y , Guan X . miR-17-5p and miR-19b-3p prevent osteoarthritis progression by targeting EZH2 . Exp Ther Med . 2020 ; 20 ( 2 ): 1653 – 1663 . Crossref PubMed Google Scholar
48. Cantley LC . The phosphoinositide 3-kinase pathway . Science . 2002 ; 296 ( 5573 ): 1655 – 1657 . Google Scholar
49. Loeser RF , Gandhi U , Long DL , Yin W , Chubinskaya S . Aging and oxidative stress reduce the response of human articular chondrocytes to insulin-like growth factor 1 and osteogenic protein 1 . Arthritis Rheumatol . 2014 ; 66 ( 8 ): 2201 – 2209 . Crossref PubMed Google Scholar
50. Yin W , Park J-I , Loeser RF . Oxidative stress inhibits insulin-like growth factor-I induction of chondrocyte proteoglycan synthesis through differential regulation of phosphatidylinositol 3-kinase-Akt and MEK-ERK MAPK signaling pathways . J Biol Chem . 2009 ; 284 ( 46 ): 31972 – 31981 . Crossref PubMed Google Scholar
51. Litherland GJ , Dixon C , Lakey RL , et al. Synergistic collagenase expression and cartilage collagenolysis are phosphatidylinositol 3-kinase/Akt signaling-dependent . J Biol Chem . 2008 ; 283 ( 21 ): 14221 – 14229 . Crossref PubMed Google Scholar
52. Mengshol JA , Vincenti MP , Brinckerhoff CE . IL-1 induces collagenase-3 (MMP-13) promoter activity in stably transfected chondrocytic cells: requirement for Runx-2 and activation by p38 MAPK and JNK pathways . Nucleic Acids Res . 2001 ; 29 ( 21 ): 4361 – 4372 . Crossref PubMed Google Scholar
Author contributions
Z. Luobu: Investigation, Supervision, Writing – original draft.
L. Wang: Methodology, Validation, Writing – original draft.
D. Jiang: Supervision, Project administration, Writing – review & editing.
T. Liao: Writing – review & editing, Validation.
C. Luobu: Formal Analysis, Supervision.
L. Qunpei: Conceptualization, Funding acquisition, Project administration, Resources.
Funding statement
No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
ICMJE COI statement
The authors declare that they have no financial conflicts of interest.
Acknowledgements
We thank the Department of Orthopedics, Lhasa People's Hospital for providing the osteoarthritis (OA) tissue samples and related anonymous clinical data.
Ethical review statement
Written informed consent was obtained from patients with approval by the Institutional Review Board in Lhasa People's Hospital.
Open access funding
The authors report that the open access funding for their manuscript was self-funded.
Supplementary material
Figure showing the effects of circSCAPER on chondrocytes without interleukin (IL)-1β.









